Abstract
The proteinase-activated receptor-2 (PAR-2) is the second member of a putative larger class of proteolytically activated receptors that mediate cell activation events by receptor cleavage or synthetic peptidomimetics corresponding to the newly generated N-terminus. To further study the previously identified mitogenic effects of PAR-2, we used the interleukin-3 (IL-3)–dependent murine lymphoid cell line, BaF3, for generation of stable cell lines expressing PAR-2 (BaF3/PAR-2) or the noncleavable PAR-2 mutant PAR-2Arg36 → Ala36. Only BaF3 cells expressing either wild-type or mutated receptor exhibited mitogenic responses when grown in IL-3–deficient media supplemented with PAR-2 activating peptide (SLIGRL, PAR39-44). This effect was dose dependent with an EC50 of ∼80 μmol/L, sustained at 24, 48, and 72 hours, and was also demonstrable using thrombin receptor peptide TR42-47. Because tryptase shares ∼70% homology with trypsin (previously shown to activate PAR-2), we studied recombinantly expressed forms of α- and β-tryptases as candidate protease agonists for PAR-2. Hydrolytic activity of the chromogenic substrate tosyl-glycyl-prolyl-argly-4-nitroanilide acetate was present as a sharp peak at Mr ∼130, confirming the presence of secretable and functionally active homotetrameric α- and β-tryptases in transfected COS-1 cells. Dose-dependent proliferative responses were evident using either secreted form of tryptase with maximal responses seen at ∼3 pmol/L (0.1 U/L). Receptor proteolysis was necessary and sufficient for mitogenesis because active site-blocked tryptase failed to induce this response, and proliferative responses were abrogated in BaF3 cells expressing PAR-2Arg36 → Ala36. These results specifically identify both forms of mast cell tryptases as serine protease agonists for PAR-2 and have implications for elucidating molecular mechanisms regulating cellular activation events mediated by proteases generated during inflammatory, fibrinolytic, or hemostatic-regulated pathways.
CELLULAR RESPONSIVENESS to extracellular stimuli classically involves receptor-ligand binding interactions with subsequent activation of a finely regulated repertoire of intracellular effector proteins and phosphorylation events. The observation that serine proteases, such as thrombin,1-5 plasmin,6 cathepsin G,7 and coagulation factors VII and X,8 display direct cellular effects, suggested that alternative mechanisms of receptor activation may exist; a concept that was reinforced with the isolation and cloning of a proteolytically activated thrombin receptor.9 Whereas the prototypic G-protein–coupled thrombin receptor has been extensively characterized,9-11 the identification of a second cleavable receptor, proteinase-activated receptor-2 (PAR-2),12 further intimated the existence of a more extended gene family of proteolytically activated receptors displaying similar mechanisms of activation. Like the thrombin receptor, PAR-2 is activated by proteolytic cleavage and by synthetic peptides corresponding to the newly generated N-terminus after receptor cleavage.12-15 Although trypsin clearly activates PAR-2,12 13 the presence of an additional physiological protease agonist(s) appeared likely, especially in view of the disparate cellular distributions of trypsin and PAR-212 and trypsin's relatively nonspecific enzymatic activity.
PAR-2 tissue-specific expression has been documented in the liver, kidney, intestine, and stomach,12 and cell-specific characterization has been described in human vascular endothelial cells, keratinocytes, and intestinal epithelial cells.14-16 Like the thrombin receptor, PAR-2 mediates proliferative responses in primary cultures of vascular endothelial cells, as determined by using PAR-2–specific peptidomimetics.14 Furthermore, functional cross-regulatory mechanisms were shown between both thrombin receptor (TR) and PAR-2 in vitro, suggesting that coexpression of both proteolytically cleaved receptors on vascular endothelial cells may represent a previously unrecognized molecular mechanism regulating hemostatic properties of the vessel wall.14 Extrapolation of these initial observations suggested that the PAR-2 protease agonist may display similar functional properties and presumably represents a circulating plasma protein zymogen.
To characterize the proliferative responses mediated through PAR-2, we have expressed proteolytically active α- and β-tryptases and established their relative roles in activating this receptor. Neither catalytically inactive tryptases nor a noncleavable PAR-2 mutant showed proliferative responses, confirming that receptor proteolysis was a necessary and sufficient prerequisite for downstream receptor-coupling events. These observations may resolve previous observations outlining stimulatory effects of mast cells17 and tryptases18 on various cell types. On a broader scale, these observations define proteolytically activated receptors as an emerging family of cell surface receptors that mediate cell activation events elicited by serine proteases generated during inflammatory, fibrinolytic, or thrombosis-regulated pathways.
MATERIALS AND METHODS
Supplies, reagents, and cell lines. Restriction enzymes were purchased from Stratagene (La Jolla, CA). The fluorescein isothiocyanate (FITC)-conjugated affinity-purified F(ab′)2 goat antirabbit antibody was purchased from Tago, Inc (Burlingame, CA). The thrombin receptor-activating peptide TR42-47(SFLLRN), the murine PAR-2–activating peptide (SLIGRL, PAR39-44), and the inactive peptide in which the first two residues were exchanged (LSIGRL), have been previously described and characterized.14 The interleukin-3 (IL-3)–dependent murine pro-B lymphoid cell line BaF3 was kindly provided by Dr J.-P. Li (New York University Medical Center). BaF3 cells were propagated in RPMI 1640 (GIBCO/BRL, Gaithersburg, MD) supplemented with 10% fetal calf serum, 10% WEHI-3 medium (as the IL-3 source), penicillin (100 U/mL), and streptomycin (100 μg/mL). COS-1 cells were grown in Dulbecco's modified Eagle's medium (DMEM; GIBCO/BRL) supplemented with 10% fetal calf serum, penicillin (100 U/mL), and streptomycin (100 μg/mL). Twice-crystallized trypsin was purchased from Sigma (St Louis, MO) and had a specific activity of 12,700 U/mg protein.
Generation of a stable BaF3/PAR-2 cell line. The human PAR-2 cDNA14 was cloned into the EcoRI site of the eukaryotic expression plasmid pcDNA3 (Stratagene) and its orientation confirmed by restriction analysis. The PAR-2 noncleavable mutant (PAR-2Arg36 → Ala36) was generated by PCR mutagenesis and fully sequenced for confirmation. BaF3 cells (3 × 107 cells/mL resuspended in HEPES-buffered saline [HBS; pH 7.4]) were transfected with 20 μg of linearized plasmids by electroporation using a Bio-Rad Gene Pulser set at 290 V and 1,980 μF (Bio-Rad, Richmond, CA). Electroporated cells were centrifuged at 1,500 revolutions per minute, washed in phosphate-buffered saline (PBS), and propagated for 48 hours in complete media (containing WEHI-3) to allow recovery. Subsequently, wild-type, pcDNA3-transfected (mock controls) or BaF3/PAR-2 cells were incubated in progressive concentrations of G418 (GIBCO/BRL) for selection of resistant clones. By using this strategy, we noted that G418 at an initial concentration of 400 μg/mL failed to induce cell death in wild-type cells, although concentrations of 800 μg/mL resulted in complete cell death. Consequently, a concentration of 1 mg/mL was used for final selection. Surviving cells were pooled and propagated for further analyses.
Cell-surface receptor expression was confirmed by using a human PAR-2–specific antibody and flow cytometric analysis. The antibody is an affinity-purified rabbit polyclonal IgG directed against residues Ser37 -Val53 of human PAR-2; its generation and characterization will be described elsewhere. Briefly, 1 × 106 cells were incubated with 20 mg/mL anti–PAR-237-53 for 45 minutes at 4°C, washed one time with PBS, and subsequently incubated with a 1:100 dilution of the FITC-conjugated secondary antibody for 45 minutes at 4°C. After extensive washing in PBS, cells were analyzed for PAR-2 expression in a fluorescence-activated cell-sorting (FACS) trianalyzer (Becton Dickinson, Mountain View, CA).
For mitogenesis assays, 2 × 104 cells were transferred to 96-well clusters (tissue-culture treated polystyrene; Falcon Labware, Lincoln Park, NJ ) and propagated in WEHI-3–deficient medium for analysis of individual agonists. Cells grown in WEHI-3–supplemented medium functioned as positive controls whereas cells propagated in WEHI-3–deficient medium consistently died within 24 hours and functioned as negative controls. Cells were then incubated for specified time periods (24, 48, or 72 hours) at 37°C and cell numbers were determined at the end of these time periods by using a tetrazolium-based colorimetric assay (MTT) for cell proliferation and survival, essentially as previously described.19
Tryptase expression and characterization. Human mast cell cDNAs for α- and β-tryptases20,21 were kindly provided by Dr L. Schwartz (Medical College of Virginia, Richmond, VA) and used for generation of eukaryotic expression plasmids encoding either α-tryptase (pcDNAα) or β-tryptase (pcDNAβ). Transient transfections in COS-1 cells were completed by using DEAE dextran. Briefly, COS-1 cells were plated at a density of 1 × 106 cells/100 mm2 plate, and the following day transfections were completed by using DMEM containing 10% heat-inactivated NuSerum, 300 μg/mL DEAE dextran, 100 μmol/L chloroquine and 10 μg of individual plasmid constructs. Transfections were allowed to proceed for 4 hours at 37°C, 5% CO2 , and cells were washed once with DMEM and incubated for 2 minutes in 10% dimethyl sulfoxide (DMSO) in PBS at 25°C. Cells were then washed and propagated in complete media for 24 hours at which point the media was replaced with 2.5 mL of complete medium supplemented with 50 μg/mL heparin, which has been shown to facilitate retention of the tryptase homotetrameric forms.18 At 72 hours, the supernatants containing secreted forms of tryptases were harvested and stored at −70°C for further use. Cells were simultaneously harvested by using a rubber policeman in 2.5 mL of a buffer containing 0.01 mol/L 2-(N-morpholino)ethane sulfonic acid (MES), pH 6.5 and 1 mol/L NaCl supplemented with 50 μg/mL heparin. Cell lysis was completed by sonication, and cellular debris separated by centrifugation at 4,000g for 40 minutes at 4°C. The pellet was discarded and the supernatant (intracellular tryptase) was stored at −70°C for further use. Protein concentration of individual sample supernatants and cell extracts was measured by the method of Bradford,22 by using bovine serum albumin (BSA) as a standard. Samples were diluted with the same buffer to adjust to uniform protein concentration.
Analytical techniques. Intracellular and secreted tryptases in conditioned media from transfected COS-1 cells were analyzed for tryptase expression by radioimmunoassay (Pharmacia AB, Uppsala, Sweden).23 Briefly, 50 μL of individual samples were diluted in diluent buffer and then incubated with the primary antitryptase antibody (G5) prebound to glass test tubes for one hour at 25°C. Subsequently, samples were incubated with the I125-labeled murine monoclonal antitryptase antibody (G4) for 18 hours at 25°C. Samples were washed three times with 0.9% NaCl, and the amount of bound radioactivity was quantitated by scintillography. Each sample was determined in duplicate and expressed against a known standard concentration of tryptase generated by standard curves (1 U/L equivalent to 1 μg purified tryptase [∼30 pmol/L]). Tryptase proteolytic activity was evaluated by using two distinct chromogenic substrates: tosyl-glycyl-prolyl-lysine-4-nitroanilide-acetate (TGPL) and tosyl-glycyl-prolyl-argyl-4-nitroanilide acetate (TGPA; Boehringer-Mannheim, Mannheim, Germany). These initial experiments established that tryptase showed enhanced hydrolytic activity using TGPA, and all subsequent experiments were carried out by using this substrate. Reactions were completed in 0.1 mol/L Tricine/NaOH (pH 8.3), 50 mmol/L NaCl, 25 mmol/L EDTA, 0.1% BSA and Antifoam C at 37°C using 0.3 mmol/L substrate. p-Nitroanilide generation was quantified at 405 nm and hydrolytic activity fitted to a standard curve using substrate cleavage by trypsin. The latter curve was essentially linear in the range 1 pmol/L to 10 nmol/L. To confirm that the hydrolytic activity comigrated with the known homotetrameric active forms of tryptase, expressed samples were subjected to gel filtration chromatography. Briefly, 400 μL of the individual samples were loaded on a Superdex 200 HR preparative grade gel filtration column (0.9 cm × 60 cm; Pharmacia LKB) pre-equilibrated in 150 mmol/L NaCl, 10 mmol/L EDTA, and 10 mmol/L sodium phosphate, pH 7.36. The column was eluted at a flow rate of 4 mL/h, and 1-mL fractions were analyzed for immunoreactive and proteolytic activity by using known protein standards for size determinations.
The inhibition profile of these recombinant tryptases to known protease inhibitors was also studied. Briefly, 50 μL of the secreted or intracellular forms of tryptases were preincubated for 15 minutes at 37°C with various inhibitors (10 μg/mL soybean trypsin inhibitor, 2 mmol/L EDTA, 10 μg/mL aprotinin, 2 mmol/L benzamidine, and 0.2 mmol/L leupeptin), and the proteolytic activity was compared with matched samples using tryptase alone as described previously.
RESULTS
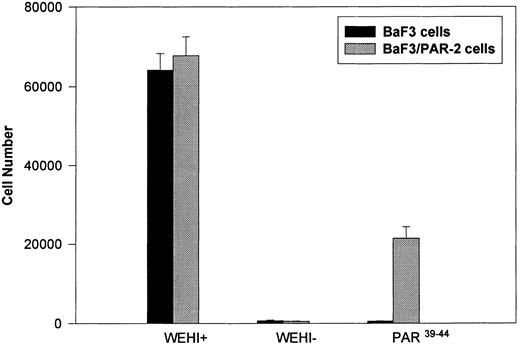
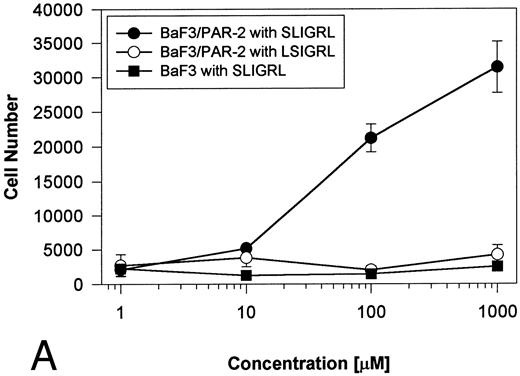
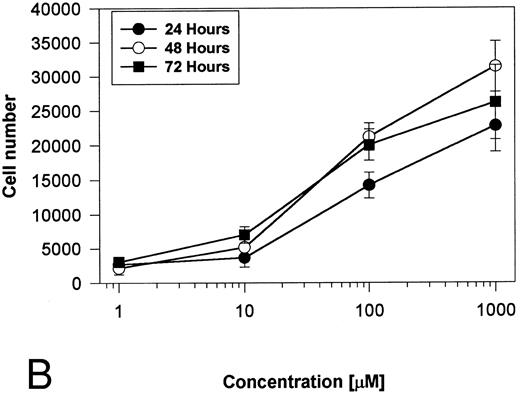
Generation and characterization of a BaF3/PAR-2 cell line. To initiate studies designed to elucidate molecular mechanisms of PAR-2–mediated mitogenesis, we established a stable cell line using BaF3 cells. These IL-3–dependent cells have been previously shown to mediate mitogenic signals to their respective ligands when transfected with the FGF receptor24 or the erythropoietin receptor,25 suggesting that they may be suitable for analysis of mitogenic properties of G protein-coupled receptors. Stable cell lines expressing PAR-2 (or mock-transfected controls) were generated by transfection of PAR-2 cDNA into BaF3 cells by electroporation. The proliferative characteristics of the transfected BaF3/PAR-2 cell line were then examined in more detail. As shown in Fig 1, both mock-transfected BaF3 and BaF3/PAR-2 cells displayed mitogenic responses to WEHI-3 medium at 48 hours, whereas cells incubated in the absence of WEHI-3 were completely apoptotic at this time point. In contrast, only the BaF3/PAR-2 cells showed proliferative responses when incubated with 100 μmol/L PAR39-44, a concentration previously shown to induce mitogenesis in human umbilical vein endothelial cells.14 These responses were nearly 25% to 30% of the maximal response observed using 10% WEHI-3 medium. BaF3/PAR-2 cells specifically showed concentration-dependent mitogenic responses to PAR39-44, with an EC50 of approximately 80 μmol/L (Fig 2A), with no response to the inactive scrambled peptide LSIGRL or mock-transfected (or wild-type) BAF3 cells. Furthermore, these proliferative responses to PAR39-44 were sustained at three different time points (Fig 2B). These initial observations established that BaF3 cells contained the requisite signaling pathways for mediating proliferation responses through this G protein-coupled receptor and that PAR-2 expression could confer IL-3 independence to this cell line when incubated with its specific ligand.
Generation and characterization of BaF3/PAR-2 cells. Mock-transfected (BaF3 cells) or BaF3/PAR-2 cells were plated into 96-well clusters (2 × 104 cells/well) and incubated in complete media (RPMI, 10% fetal calf serum) with (WEHI+) or without (WEHI−) 10% WEHI supernatant (as the exogenous source of IL-3), or 100 μmol/L PAR39-44 for 48 hours, before proliferative assays using MTT essentially as outlined in the Materials and Methods. Only BaF3/PAR-2 cells showed proliferative responses to PAR-2 peptidomimetics. No responses were seen using the inactive peptide LSIGRL (not shown). All points represent the mean ± the standard error of the mean (SEM) of four wells from a single representative set of experiments.
Generation and characterization of BaF3/PAR-2 cells. Mock-transfected (BaF3 cells) or BaF3/PAR-2 cells were plated into 96-well clusters (2 × 104 cells/well) and incubated in complete media (RPMI, 10% fetal calf serum) with (WEHI+) or without (WEHI−) 10% WEHI supernatant (as the exogenous source of IL-3), or 100 μmol/L PAR39-44 for 48 hours, before proliferative assays using MTT essentially as outlined in the Materials and Methods. Only BaF3/PAR-2 cells showed proliferative responses to PAR-2 peptidomimetics. No responses were seen using the inactive peptide LSIGRL (not shown). All points represent the mean ± the standard error of the mean (SEM) of four wells from a single representative set of experiments.
Time- and concentration-dependent responses to PAR39-44. (A) Mock-transfected BaF3 cells (open circles) or BaF3/PAR-2 cells (closed circles) were plated as outlined in the legend to Fig 1 and incubated with progressive concentrations of PAR39-44 or SLIGRL (closed squares) for 48 hours before MTT analysis. (B) BaF3/PAR-2 cells were studied in the identical fashion for variable periods of time as delineated. All experiments were completed in WEHI− media. Results are expressed as the mean ± SEM of four wells from a single set of experiments.
Time- and concentration-dependent responses to PAR39-44. (A) Mock-transfected BaF3 cells (open circles) or BaF3/PAR-2 cells (closed circles) were plated as outlined in the legend to Fig 1 and incubated with progressive concentrations of PAR39-44 or SLIGRL (closed squares) for 48 hours before MTT analysis. (B) BaF3/PAR-2 cells were studied in the identical fashion for variable periods of time as delineated. All experiments were completed in WEHI− media. Results are expressed as the mean ± SEM of four wells from a single set of experiments.
Expressed forms of tryptases are proteolytically active in eukaryotic cells. Because tryptase is a “trypsin-like” serine protease with previously demonstrated mitogenic potential,18 we specifically investigated whether expressed forms of human mast cell α- or β-tryptase could induce proliferation through PAR-2. Eukaryotic expression plasmids for α- or β-tryptase were constructed and used for transient transfection assays in COS-1 cells. Expression levels were quantified in supernatant conditioned media and cell extracts were obtained at 72 hours after transfection with a radioimmunoassay. As shown in Table 1, quantifiable measurements of α- and β-tryptase were found in cell extracts (intracellular form) and supernatants (secreted forms) with quantifications well within the range for detection using the radioimmunoassay.23 There was no evidence for tryptase in control mock-transfected cells. Confirmation that the recombinant tryptases retained proteolytic activity was shown by using the known chromogenic substrates for tryptase and trypsin, TGPL and TGPA. TGPA functioned as a more active tryptase substrate than TGPL in preliminary analyses using both α- and β-tryptase (data not shown) and was used for comparative determination of proteolytic activity, using trypsin as a standard. As outlined in Table 1, both secreted and intracellular forms of α-tryptase showed hydrolytic activity, approximating 8 pmol/L and 1 pmol/L trypsin equivalents/U tryptase. In contrast, only the secreted form of β-tryptase displayed proteolytic activity, with no activity detected using the intracellular (non-secreted) form of the recombinant protein (see Discussion).
Characterization of Recombinant Expressed Tryptases in COS-1 Cells
| . | Secreted Forms . | Intracellular Forms . | ||
|---|---|---|---|---|
| . | Concentration* . | Proteolytic Activity† . | Concentration . | Proteolytic Activity . |
| α-Tryptase | 9 | 8.3 | 11 | 1.0 |
| β-Tryptase | 64 | 39.1 | 38 | <0.001 |
| Mock-transfected | <0.5 | <0.001 | <0.5 | <0.001 |
| . | Secreted Forms . | Intracellular Forms . | ||
|---|---|---|---|---|
| . | Concentration* . | Proteolytic Activity† . | Concentration . | Proteolytic Activity . |
| α-Tryptase | 9 | 8.3 | 11 | 1.0 |
| β-Tryptase | 64 | 39.1 | 38 | <0.001 |
| Mock-transfected | <0.5 | <0.001 | <0.5 | <0.001 |
COS-1 cells were transfected with pcDNAα (α-tryptase), pcDNAβ (β-tryptase), or pcDNA alone (mock-transfected), and supernatants and cell extracts were harvested at 72 hours (48 hours in the presence of 50 μg/mL heparin) as outlined in Materials and Methods. Samples were analyzed for tryptase expression using radioimmunoassay (RIA) and represent the mean from duplicate determinations, expressed in units per liter, standardized to cell number as determined by protein quantification.
Proteolytic activity was determined using the chromogenic substrate TGPA as outlined in the Materials and Methods, and results are expressed in picomolar trypsin equivalents per unit of tryptase.
Proteolytically active forms of tryptase require homotetramerization (Mr of monomer ∼28). To show that the tryptic activity was found in this form, samples were subjected to gel filtration chromatography, and specific fractions assayed for proteolytic activity. As shown in Fig 3, a single peak of hydrolytic activity with an apparent Mr ∼130 was clearly evident using a secreted form of β-tryptase consistent with the presence of expressed tryptase in homotetrameric form. Less pronounced, but detectable peaks of activity at Mr ∼130 were evident with secreted and intracellular forms of α-tryptase (data not shown). Likewise, no proteolytic activity was evident in samples with Mr less than 50, all consistent with known biochemical properties of purified tryptases.18 Finally, these recombinant tryptases were subjected to inhibition profile analysis using a variety of protease inhibitors. As shown in Table 2, tryptic activity in samples was essentially abrogated by preincubation with benzamidine and leupeptin, whereas EDTA and soybean trypsin inhibitor were unable to significantly suppress tryptic activity. This pattern is essentially identical to the previously described inhibition profile of human lung and dog mastocytoma tryptase,26 confirming that both forms of proteolytically active tryptase could be expressed in eukaryotic cells retaining biochemical properties of previously purified tryptases.
Gel-filtration chromatography using β-tryptase supernatant. A total of 400 mL of individual samples (α- and β-tryptase intracellular or secreted forms) were loaded on a Superdex 200 HR preparative grade gel filtration column, and 1-mL fractions were collected and analyzed for tryptase enzymatic activity as described in the Materials and Methods. The elution profile of three reference protein standards (BSA [67 kD], aldolase [158 kD], and catalase [232 kD]) is depicted. A single, maximal peak of hydrolytic activity comigrating with the known homotetrameric form of β-tryptase (∼130 kD) is evident, with no hydrolytic activity seen at other molecular weights. Identical (but smaller) peaks of proteolytic activity were seen using both secreted and intracellular forms of α-tryptase but not intracellular β-tryptase (not shown).
Gel-filtration chromatography using β-tryptase supernatant. A total of 400 mL of individual samples (α- and β-tryptase intracellular or secreted forms) were loaded on a Superdex 200 HR preparative grade gel filtration column, and 1-mL fractions were collected and analyzed for tryptase enzymatic activity as described in the Materials and Methods. The elution profile of three reference protein standards (BSA [67 kD], aldolase [158 kD], and catalase [232 kD]) is depicted. A single, maximal peak of hydrolytic activity comigrating with the known homotetrameric form of β-tryptase (∼130 kD) is evident, with no hydrolytic activity seen at other molecular weights. Identical (but smaller) peaks of proteolytic activity were seen using both secreted and intracellular forms of α-tryptase but not intracellular β-tryptase (not shown).
Inhibition Profile of α- and β-Tryptases
| Protease Inhibitor . | Percent Control . | ||
|---|---|---|---|
| . | β-Tryptase . | α-Tryptase . | α-Tryptase (Intracellular) . |
| . | (Secreted) . | (Secreted) . | . |
| — | 100 | 100 | 100 |
| EDTA (2 mmol/L) | 95 ± 0.5 | 92 ± 2.5 | 91 ± 1.6 |
| Soybean trypsin inhibitor (10 μg/mL) | 82 ± 0.2 | 85 ± 1.5 | 78 ± 2.8 |
| Aprotinin (10 μg/mL) | 30 ± 0.6 | 35 ± 2.5 | 33 ± 1.8 |
| Benzamidine (2 mmol/L) | 3.2 ± 0.5 | 4.5 ± 1.0 | 3.5 ± 1.2 |
| Leupeptin (0.2 mmol/L) | 1.5 ± 0.3 | 1.0 ± 0.4 | 1.5 ± 0.5 |
| Protease Inhibitor . | Percent Control . | ||
|---|---|---|---|
| . | β-Tryptase . | α-Tryptase . | α-Tryptase (Intracellular) . |
| . | (Secreted) . | (Secreted) . | . |
| — | 100 | 100 | 100 |
| EDTA (2 mmol/L) | 95 ± 0.5 | 92 ± 2.5 | 91 ± 1.6 |
| Soybean trypsin inhibitor (10 μg/mL) | 82 ± 0.2 | 85 ± 1.5 | 78 ± 2.8 |
| Aprotinin (10 μg/mL) | 30 ± 0.6 | 35 ± 2.5 | 33 ± 1.8 |
| Benzamidine (2 mmol/L) | 3.2 ± 0.5 | 4.5 ± 1.0 | 3.5 ± 1.2 |
| Leupeptin (0.2 mmol/L) | 1.5 ± 0.3 | 1.0 ± 0.4 | 1.5 ± 0.5 |
A total of 50 μL of the active recombinant tryptases were incubated with various known protease inhibitors for 15 minutes at 37°C. After incubation, proteolytic activity of the mixtures was compared to tryptase alone using TGPA and p-Nitroanilide generation determined at 405 nm. Activities are expressed as a percentage of maximum.
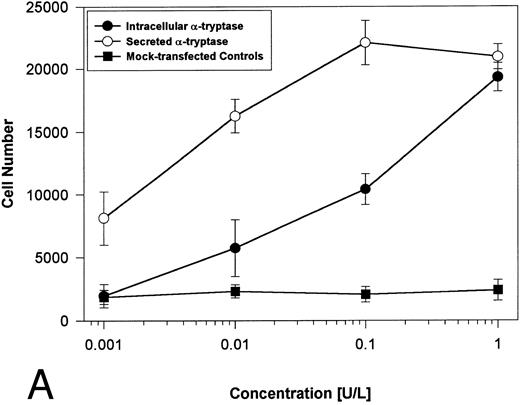
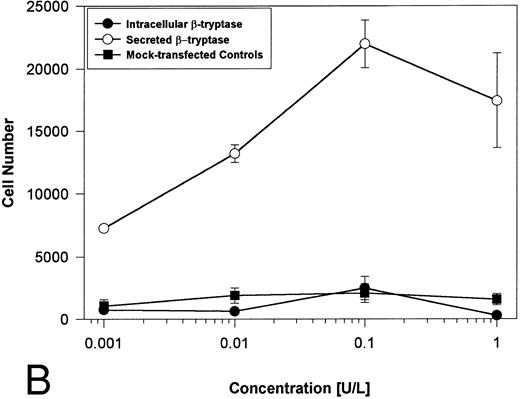
Proteolytically active tryptases induce proliferation through PAR-2. The ability of these expressed forms of α- or β-tryptases to induce proliferation in BaF3/PAR-2 cell lines was then studied. Cells were incubated with known quantities of the individual tryptases and mitogenic responses studied at 48 hours. As shown in Fig 4A, both secreted and intracellular forms of α-tryptase elicited a concentration-dependent proliferative response in BaF3/PAR-2 cells, with no responses evident in mock-transfected controls. Proliferative responses using secreted forms of β-tryptase were essentially identical to those seen using secreted α-tryptase, showing nearly superimposable concentration-dependent response curves (Fig 4B). The estimated EC50 using either secreted forms of α- or β-tryptase approximated 0.1 U/L (0.3 pmol/L). When compared with control wells activated with 100 μmol/L PAR39-44, peak proliferative responses using either of the secreted tryptases (∼0.1 U/L) were approximately 75% of those seen with PAR39-44. No proliferative response to either α- or β-tryptase was evident in mock-transfected BaF3 cells acting as controls or in mock-transfected COS-1 supernatants or cellular extracts (data not shown). In contrast to the responses outlined above, nonsecreted (proteolytically inactive) β-tryptase failed to elicit any proliferative response in BaF3/PAR-2 cell lines, even at concentrations up to 1 U/L, consistent with a requirement for tryptase hydrolytic activity.
Proliferative responses of BaF3/PAR-2 cells to α- (A) or β-tryptases (B). BaF3/PAR-2 cells were plated as outlined in the legend to Fig 1 and incubated with variable amounts of α- or β-tryptases (in the absence of WEHI supplement) for 48 hours before functional evaluation (1 U/L tryptase is equivalent to ∼30 pmol/L). No proliferative responses were evident using mock-transfected BaF3 cells. Likewise, mock-transfected COS-1 supernatants or extracts diluted 1:1 vol/vol with BaF3/PAR-2 cells failed to show any proliferation (not shown). Results represent the mean ± SEM of four wells from a representative set of experiments.
Proliferative responses of BaF3/PAR-2 cells to α- (A) or β-tryptases (B). BaF3/PAR-2 cells were plated as outlined in the legend to Fig 1 and incubated with variable amounts of α- or β-tryptases (in the absence of WEHI supplement) for 48 hours before functional evaluation (1 U/L tryptase is equivalent to ∼30 pmol/L). No proliferative responses were evident using mock-transfected BaF3 cells. Likewise, mock-transfected COS-1 supernatants or extracts diluted 1:1 vol/vol with BaF3/PAR-2 cells failed to show any proliferation (not shown). Results represent the mean ± SEM of four wells from a representative set of experiments.
This apparent requirement for proteolytically active tryptase was directly addressed by reassessing mitogenic responses using secreted β-tryptase blocked with various protease inhibitors (see Table 2). BaF3/PAR-2 cells were incubated with β-tryptase alone or with β-tryptase preincubated with various protease inhibitors, and the effects on proliferative potential were further analyzed. As shown in Fig 5, only leupeptin and benzamidine (protease inhibitors that had both shown significant capacity in abrogating tryptase proteolytic activity) were able to significantly inhibit tryptase-induced mitogenic responses in these cells. This observation reinforces the concept that the mitogenic effects mediated through PAR-2 require receptor cleavage by proteolytically active tryptase.
Effect of protease inhibitors on tryptase-induced proliferative responses. BaF3/PAR-2 cells were plated as outlined in the legend to Fig 1 and incubated for 48 hours with 0.1 U/L (3 pmol/L) β-tryptase or β-tryptase that had been preincubated with specified protease inhibitors for 15 minutes at 37°C. Inhibitor concentrations were 2 mmol/L EDTA, 10 μg/mL soybean trypsin inhibitor (STI), 2 mmol/L benzamidine (BZA), and 0.2 mmol/L leupeptin. As shown, tryptase-mediated proliferative responses are specifically abrogated by protease inhibitors previously shown to block tryptase hydrolytic activity (see Table 2). Results represent the mean ± SEM of four wells from a single representative set of experiments.
Effect of protease inhibitors on tryptase-induced proliferative responses. BaF3/PAR-2 cells were plated as outlined in the legend to Fig 1 and incubated for 48 hours with 0.1 U/L (3 pmol/L) β-tryptase or β-tryptase that had been preincubated with specified protease inhibitors for 15 minutes at 37°C. Inhibitor concentrations were 2 mmol/L EDTA, 10 μg/mL soybean trypsin inhibitor (STI), 2 mmol/L benzamidine (BZA), and 0.2 mmol/L leupeptin. As shown, tryptase-mediated proliferative responses are specifically abrogated by protease inhibitors previously shown to block tryptase hydrolytic activity (see Table 2). Results represent the mean ± SEM of four wells from a single representative set of experiments.
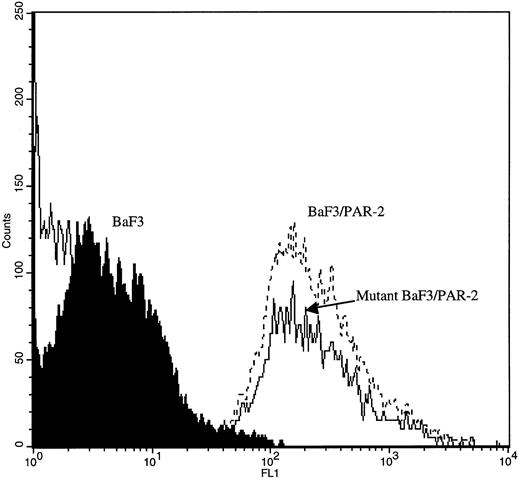
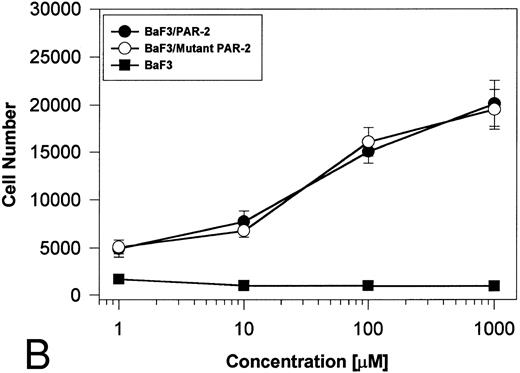
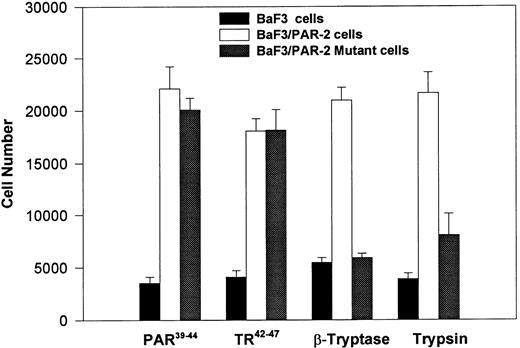
PAR-2 cleavage is necessary for mitogenesis. The studies outlined previously were consistent with a receptor cleavage mechanism as a requisite step in inducing mitogenic potential through PAR-2. This hypothesis was subsequently confirmed by generation of a noncleavable PAR-2 mutant in which the active site arginine was mutated to alanine (Arg36 → Ala36 ). As shown by flow cytometric analysis using an anti–PAR-2 antibody, similar levels of receptor expression were evident in stably transfected BaF3/PAR-2 cells or mutant BaF3/PAR-2 cells (Fig 6). Likewise, both cell lines showed similar proliferative characteristics when activated by PAR-2–specific peptide ligands (Fig 7A). Because recent data suggested that thrombin receptor peptidomimetics could also activate PAR-2,27 we compared proliferative characteristics of these cells to TR42-47. As can be seen in Fig 7B, cell lines expressing either receptor showed comparable proliferative responses by using either synthetic peptide ligand, although proliferative responses using TR42-47 were approximately 70% of those seen using equimolar concentrations of PAR-2 peptides. These proliferative responses mediated by synthetic peptidomimetics are consistent with previously described thrombin receptor structure/function studies, in which the peptide ligand binding site represents an intramolecular pocket independent of the receptor cleavage site. Proliferative responses of both cell lines were then studied by using β-tryptase. As can be seen in Fig 8, mutant PAR-2 cells incubated with proteolytically active tryptase failed to proliferate at concentrations as high as 1 U/L (30 pmol/L), concentrations that were clearly saturating when used in BaF3/PAR-2 cells. Likewise, only cells transfected with PAR-2Arg36 → Ala36 failed to proliferate when activated by trypsin, although BaF3/PAR-2 cells showed proliferative responses not unlike those seen when using saturating concentrations of tryptase. Thus, neither proteolytically inactive tryptases nor noncleavable PAR-2 are able to mediate proliferative signals, showing that receptor proteolysis is necessary and sufficient for protease-induced cell activation events.
Flow cytometric analysis in transfected cell lines. Mock-transfected BaF3, BaF3/PAR-2, or BaF3/PAR-2Arg36 → Ala36 (mutant) cells were incubated with 20 μg/mL anti–PAR-2 antibody for flow cytometric analysis using a FITC-conjugated secondary antibody, as outlined in the Materials and Methods. As shown, comparable cell-surface receptor expression is seen in both cell lines expressing PAR-2 (<95% immunoreactive cells), whereas no immunoreactivity is detectable using mock-transfected BaF3 controls.
Flow cytometric analysis in transfected cell lines. Mock-transfected BaF3, BaF3/PAR-2, or BaF3/PAR-2Arg36 → Ala36 (mutant) cells were incubated with 20 μg/mL anti–PAR-2 antibody for flow cytometric analysis using a FITC-conjugated secondary antibody, as outlined in the Materials and Methods. As shown, comparable cell-surface receptor expression is seen in both cell lines expressing PAR-2 (<95% immunoreactive cells), whereas no immunoreactivity is detectable using mock-transfected BaF3 controls.
PAR-2 proliferative responses to synthetic peptidomimetics. BaF3/PAR-2 or BaF3/PAR-2Arg36 → Ala36 (mutant) cell lines were incubated with progressive concentrations of PAR-2–derived (PAR-239-44) (A) or TR-derived (TR42-47) (B) peptidomimetics, essentially as described in the legend to Fig 1. As shown, both cell lines responded almost identically when activated by either TR42-47 or PAR-239-44, although the responses using TR42-47 were consistently ∼70% of those seen using equimolar concentrations of PAR39-44. Results represent the mean ± SEM from four individual wells from a representative set of experiments completed on two occasions.
PAR-2 proliferative responses to synthetic peptidomimetics. BaF3/PAR-2 or BaF3/PAR-2Arg36 → Ala36 (mutant) cell lines were incubated with progressive concentrations of PAR-2–derived (PAR-239-44) (A) or TR-derived (TR42-47) (B) peptidomimetics, essentially as described in the legend to Fig 1. As shown, both cell lines responded almost identically when activated by either TR42-47 or PAR-239-44, although the responses using TR42-47 were consistently ∼70% of those seen using equimolar concentrations of PAR39-44. Results represent the mean ± SEM from four individual wells from a representative set of experiments completed on two occasions.
PAR-2 proliferative responses require receptor cleavage. Individual cell lines were incubated with 100 μmol/L PAR-239-44, 100 μmol/L TR42-47, 0.1 U/L tryptase or 1 nmol/L trypsin, and analyzed for proliferative responses at 48 hours. As shown, proliferative responses are abolished using the noncleavable PAR-2 mutant, when activated by either trypsin or β-tryptase. Results represent the mean ± SEM from six wells from a single set of experiments repeated on one occasion.
PAR-2 proliferative responses require receptor cleavage. Individual cell lines were incubated with 100 μmol/L PAR-239-44, 100 μmol/L TR42-47, 0.1 U/L tryptase or 1 nmol/L trypsin, and analyzed for proliferative responses at 48 hours. As shown, proliferative responses are abolished using the noncleavable PAR-2 mutant, when activated by either trypsin or β-tryptase. Results represent the mean ± SEM from six wells from a single set of experiments repeated on one occasion.
DISCUSSION
The concept that serine proteases may display profound cellular effects has engendered an emerging paradigm focusing on the molecular mechanisms of cellular/protease interactions. Previous data suggest that some of these cellular responses are mediated by an emerging class of G-protein–coupled proteolytically activated receptors (PARs) exemplified by the thrombin receptor and most recently the PAR-2. Both seven-transmembrane segment cell surface receptors display unique (but similar) modes of activation, mediate proliferative signals on activation by their respective peptide ligands, and are encoded by similarly organized genes that cocluster within the same region of human chromosome 5q13.13,28-30 Previous work in this laboratory had shown PAR-2 expression by Northern and functional analysis in vascular endothelial cells, suggesting that a PAR-2 protease may represent a naturally circulating plasma protein.14 Although trypsin clearly activates PAR-2, the expression of PAR-2 on vascular endothelial cells, coupled with trypsin's limited tissue distribution and nonspecific substrate cleavage, suggested the existence of an additional PAR-2–specific protease agonist. These initial observations further suggested that this protease would circulate as an inactive zymogen, presumably activated as one component of the three major protease-generating mechanisms in humans: fibrinolytic, hemostatic, or inflammatory pathways.
Because mast cell tryptases and trypsin share significant homology within their catalytic regions,21 and tryptase is known to be mitogenic for a number of different cell types,31-33 we used the previously described mitogenic properties of PAR-2 to delineate the role of these tryptases in activating PAR-2. Because most preparations of tissue-purified tryptases contain both α and β forms, we decided to express and characterize the role of individual tryptases in activating PAR-2. Expressed forms of both α- or β-tryptase retained proteolytic activity and showed biochemical properties consistent with those of purified tissue tryptases. To our knowledge this is the first evidence that recombinant tryptases expressed in mammalian cells retain functional activity. Both forms of tryptase induced proliferative responses only in cells expressing PAR-2, although nonsecreted (intracellular) β-tryptase was clearly nonfunctional (see below). These proliferative responses appeared optimal at tryptase concentrations of 0.1 U/L (∼3 pmol/L), although they were evident at concentrations as low as 0.03 pmol/L. The RIA used for tryptase quantification may have an ∼10-fold enhanced sensitivity for β-tryptase compared to α-tryptase34; however, this suggested that the EC50 for α-tryptase–induced mitogenesis could be overestimated by this degree.
The functional qualities of the individual tryptases shown in this report conform to recent data outlining the unique (and divergent) processing pathways for mast cell α- and β-tryptases. α-Tryptase is constitutively secreted from mast cells, and represents the predominant form normally present in the serum of healthy subjects, whereas β-tryptase is the predominant form after severe anaphylaxis and in tissue mast cells.34 The concentrations of tryptase in anaphylaxis may temporarily reach ∼2.5 nmol/L, although levels in healthy adults are not optimally quantified because of the limitations of the RIA.23,35 Unlike α-tryptase, β-tryptase requires a heparin-dependent autocatalytic processing pathway for activation. According to the proposed model,36 β-protryptase is processed at acid pH in the presence of heparin by an autocatalytic cleavage at Arg−3/Val−2 with subsequent removal of the dipeptide by dipeptidyl peptidase. Our data confirm that COS-1 cells (when supplemented with exogenous heparin) are fully able to process and secrete activated forms of β-tryptase. The inability of the intracellular forms of β-tryptase to induce proliferation of BaF3/PAR-2 cells presumably reflects the incomplete processing of this form, a step that is not required for α-tryptase. Furthermore, extrapolation of these observations suggests that these mitogenic responses require PAR-2 proteolysis because only the intracellular (proteolytically inactive) β-tryptase failed to induce the mitogenic repertoire. This initial impression was confirmed by two complementary approaches: (1) tryptase active-site inhibition essentially abrogated the proliferative responses, and (2) a noncleavable PAR-2 mutant failed to show proliferative potential when activated by either proteolytically active tryptase or trypsin.
The delineation of both α- and β-tryptase as physiological serine protease agonists for PAR-2 fulfills our initial postulate that the protease may be generated during inflammatory triggers. Previous data showed that tryptase possesses mitogenic potential in epithelial cells,31 airway smooth muscle cells,32 and fibroblasts,33 although its cell-surface receptor remained uncharacterized. Because these studies were completed with total cellular tryptases, the specific contributions of either α- or β-tryptase in mitogenesis could not be delineated. Likewise, although the EC50 for mitogenesis in these various studies approximated 2 nmol/L, direct comparisons are limited by lack of information on receptor quantifications and the role, if any, that purification methods have on enzymatic specific activities. Our data also refine previous observations on mast cell/cellular interactions. Tryptase is the most abundant protein component of human mast cells, because it is stored in secretory granules with the potential of full enzymatic activity.18 On activation, mast cells degranulate and release tryptase (along with heparin and histamine) into the circulation37 with evidence that the mast cell “releasate” directly stimulates endothelial cell activation.38 Indeed, recent data confirm that tryptase activates PAR-2 on human vascular endothelial cells, although a direct proliferative response was not shown.39 Although we have unequivocally shown that either α- or β-tryptases induce mitogenesis through PAR-2, we cannot exclude the presence of other PAR-2–specific serine protease agonists. Conversely, it remains unclear whether tryptase's known cellular effects are solely recapitulated via PAR-2 activation. In the case of the thrombin receptor, for example, proliferative responses on neuronal cells are reproduced by the serine protease granzyme A,40 and activation is modulated by other serine proteases such as cathepsin G7 and plasmin.41
The previously described functional coupling in vascular endothelial cells between the thrombin receptor and PAR-2 suggested the presence of an additional regulatory pathway controlling thrombin-mediated endothelial cell activation events. Indeed, this initial observation may explain recent data showing that mast cell tryptase modulates mitogenic potential of thrombin on vascular smooth muscle cells.42 Likewise, tryptase displays a number of biological properties relevant to modulation of the hemostatic response, including the degradation of procoagulant proteins and inactivation of fibrinogen, thereby affecting its availability as a thrombin substrate.43 The delineation of mast cell tryptases as physiological activators of PAR-2 extend our initial observations, and provide an additional link between hemostatic and inflammatory pathways. Thus, the presence of both tryptase and thrombin at sites of vascular injury provides not only a regulatory mechanism for procoagulant activity but also a means of regulating cell activation events to proteases generated at the cell surface during hemostasis, inflammation, and wound repair.
ACKNOWLEDGMENT
The authors thank Ms Shirley Murray for assistance with the preparation of this manuscript.
Supported by grants from the American Heart Association, New York State Affiliate and the National Institutes of Health (No. HL49141).
Address reprint requests to Wadie F. Bahou, MD, Division of Hematology, Health Sciences Center T15-040, State University of New York, Stony Brook, NY 11794-8151.

![Fig. 3. Gel-filtration chromatography using β-tryptase supernatant. A total of 400 mL of individual samples (α- and β-tryptase intracellular or secreted forms) were loaded on a Superdex 200 HR preparative grade gel filtration column, and 1-mL fractions were collected and analyzed for tryptase enzymatic activity as described in the Materials and Methods. The elution profile of three reference protein standards (BSA [67 kD], aldolase [158 kD], and catalase [232 kD]) is depicted. A single, maximal peak of hydrolytic activity comigrating with the known homotetrameric form of β-tryptase (∼130 kD) is evident, with no hydrolytic activity seen at other molecular weights. Identical (but smaller) peaks of proteolytic activity were seen using both secreted and intracellular forms of α-tryptase but not intracellular β-tryptase (not shown).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/10/10.1182_blood.v90.10.3914/3/m_bl_0045f3.jpeg?Expires=1769136030&Signature=3mWJ06x6-LcCQqobPdQjaWb~eHXzxxgPSNIJ5f0hPJEBMPSIlV12EUIhrQAHAmhsAGn6-HNd1BKY0wRH7GLgMbWuMxMXKmFaoebcvyanZX7hCZCNcxznYQwzlIfby81OOizfdkAgvRH1Q8CQPfO4pg3kmdlyqUrE8vOlJCFd17vw3Lsg-feqg2aezeKlwAUwq6TArE78sQ4DjVqQVFCUNxZv7FrXla7WvYWndmifBA3PUDxPM17RIelAcAIH6rKu8zsKWQIY-aAw75gpDd8bC0~uGSEcv~E1abDWGVp0stgE~L0CsJuzG9RzoZyzPq2OMrJAfqBOB~ITN3O0gniDqQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal