Abstract
A large English pedigree in which heterocellular hereditary persistence of fetal hemoglobin (HPFH) segregates is described. β-globin cluster deletions and γ gene promoter mutations associated with HPFH have been excluded. Of particular importance in this pedigree is the absence of any cosegregating hemoglobinopathy, thus allowing observation of the segregation pattern of this form of HPFH without the complicating effect of a β-globin gene mutation. Information gained in this study confirms that the extent of elevation of hemoglobin (Hb) F and F cells varies between affected individuals. There are one example of incomplete penetrance and three examples of father-to-son transmission, thus excluding X-linked inheritance. Consistent with previous reports, the most likely mode of inheritance is autosomal codominant. Linkage studies using a β-globin cluster microsatellite show no evidence of linkage to this chromosomal region implicating the presence of trans-acting regulatory factor(s). We have recently mapped one such locus to the chromosome 6q region in a very large Asian-Indian pedigree. Linkage to chromosome 6q in the English pedigree was excluded, thus indicating the presence of genetic heterogeneity in heterocellular HPFH.
THE PHYSIOLOGIC SWITCH from the production of fetal hemoglobin (Hb F ) to the adult form of Hb (Hb A) is usually accomplished by 2 years of age. Inherited conditions in which the level of Hb F remains above the normal adult level (<1%) with normal red blood cell indices and morphology are referred to as hereditary persistence of fetal hemoglobin (HPFH).1 The pancellular forms due to major deletions of the β-globin gene cluster and those associated with promoter mutations in the γ-globin genes are characterized by clearly increased levels of Hb F in heterozygotes and demonstrate a mendelian inheritance as alleles of the β-globin complex on chromosome 11p. However, there is another group characterized by modest elevations of Hb F levels (1% to 4%) distributed in an uneven fashion among the F cells (subset of erythrocytes containing Hb F ). In this group of HPFH cases (heterocellular HPFH ), no mutations are identifiable within the β-globin cluster, and in many cases the determinant is not linked to the β complex, implicating the presence of trans-acting factor(s).2-5 Surveys show that the distribution of F-cell values is skewed to the right and that approximately 10% of the normal population have at least 4.5% F cells.6-8 The importance of this condition is clearly demonstrated by the striking amelioration of the phenotype of individuals homozygous for β-thalassemia or sickle cell disease who also coinherit a HPFH determinant.9 10 Furthermore, the eventual characterization of the genetic basis for this form of heterocellular HPFH will provide important insights into developmentally regulated gene expression, and may lead to new therapeutic strategies for the hemoglobinopathies.
Recently, considerable progress has been made in the mapping of heterocellular HPFH determinants by linkage analysis using DNA polymorphisms. One locus has been mapped by our group to an 11-cM interval at the q22.3-q23.1 region in chromosome 6 in a very large Asian-Indian family.11 Another locus associated with variation in F-cell levels in sickle cell disease and normal adults has been mapped to the Xp22.2-p23.3 region.12 In the Asian-Indian family, regression analysis indicates that 90% of the variation in F-cell levels is accounted for by three genetic determinants: the 6q linked gene, β-thalassemia, and XmnI polymorphism in the Gγ gene promoter.11
We have investigated an extensive English family with heterocellular HPFH in which the HPFH determinant is inherited without a hemoglobinopathy, thus allowing a clearer impression of the segregation pattern for this type of HPFH. The pattern of transmission of the HPFH trait is strongly suggestive of autosomal codominant inheritance; X-linked inheritance is unlikely, considering that father-to-son transmission has occurred.
Linkage with the β-globin cluster is excluded (lod score < −2) at θ less than or equal to .01, and multipoint linkage analysis provides no evidence for linkage to the candidate 6q region. The data indicate that there is genetic heterogeneity in the unlinked HPFH phenotype, and that in addition to the 6q gene, there must be at least one other trans-acting autosomal locus that can exert an influence on Hb F levels in adults.
SUBJECTS AND METHODS
Hematology.Blood samples were collected (with informed consent) in EDTA as anticoagulant, and full blood cell counts were obtained using an automated cell counter. The percentage of Hb A2 was measured by elution and spectrophotometry after cellulose acetate electrophoresis at pH 8.9, and Hb F by alkaline denaturation.
F-cell assays were performed in peripheral blood using a monoclonal mouse anti–γ-globin chain antibody by microscopy (2 × 103 red blood cells counted per blood smear) and by fluorescence-activated cell sorting (FACS) (104 cells counted per assay).13
DNA analysis.DNA was extracted from peripheral blood leukocytes and analyzed for seven restriction fragment length polymorphisms in the β-globin gene cluster, and the β-haplotypes were derived.14 The T-C polymorphism at position −158 of the Gγ-globin gene15 was determined by XmnI restriction analysis of the 5′ region of the Gγ-globin gene amplified by polymerase chain reaction (PCR).16
PCR was used to specifically amplify the Gγ and Aγ promoter regions from +50 to −650 relative to the mRNA cap sites, and the regions were directly sequenced as previously described.16
Genotyping.The family was genotyped for 11 microsatellites (D6S408, D6S407, D6S262, D6S435, D6S457, D6S413, D6S472, D6S975, D6S976, D6S270, and D6S292) in the 6q22-q23 region11 and for a microsatellite between the δ- and β-globin genes within the β-globin cluster on chromosome 11p.17 Microsatellites were amplified by PCR and analyzed on denaturing 7 mol/L urea 6% polyacrylamide gels. PCR conditions were as follows: 94°C (4 minutes) followed by 35 cycles of 94°C (1 minute), 55°C (45 seconds), and 72°C (45 seconds), and a final extension at 72°C for 2 minutes. The separated PCR products were transferred onto positively charged nylon membranes (Hybond N+; Amersham, Amersham, UK) and hybridized with either radiolabeled (CA)n or PCR primers. These oligonucleotides were labeled by 3′ end–tailing with α32P-dCTP using calf thymus deoxynucleotidyl terminal transferase and the terminal transferase kit from Boehringer Mannheim (Germany). Hybridizations were performed at 42°C for 3 hours in 7% polyethylene glycol (PEG 6000 or 8000) and 10% sodium dodecyl sulfate (SDS). Membranes were washed once in 2× SSC and 0.1% SDS for 10 minutes at room temperature.
Confirmation of family relationships.The genetic relationships of the kindred were investigated using a panel of probes known to be specific for hypervariable regions of human DNA. Genomic DNA was completely digested with HinfI and Southern blot hybridized with seven minisatellite probes (MS1, MS2, MS29, MS31, MS43, MS51, and pλG3) labeled with 32P-CTP by random priming.18 Nonpaternity was excluded in all cases.
Linkage analysis.Two-point lod scores were calculated using the MLINK program of the LINKAGE package.19 Linkage calculations were performed under the assumption of autosomal dominant inheritance. Individuals were assumed to be in the genotypic classes AA, Aa, or aa, where the allele A is responsible for high values of F cells (≥8%). A disease gene frequency of 0.01 was used in the analysis, which takes into account the stringent criteria used in assignation of the high-F phenotype. Hardy-Weinberg equilibrium has been assumed. Three liability classes were designated: class 1, individuals with the AA or Aa genotype were assumed to display the phenotype in all cases; class 2, the assumption was made that individuals of the Aa genotype have a probability of .95 of displaying the phenotype; and class 3, individuals were assumed to be affected and to have the AA genotype. Liability class 1 was assigned to all affected individuals (except III-13, III-14, and III-15, described later) and all unaffected marriage partners. Individuals (other than marriage partners) who were coded as unaffected were assigned to liability class 2 to allow for the fact that they may in fact be of the Aa genotype but do not display the phenotype due to incomplete penetrance. The proband (III-13) and her siblings (III-14 and III-15), who had an increased level of F cells severalfold that of their parents who were both affected, were assigned to liability class 3. Analyses were made separately in both sides of the extended family, pedigrees A and B, as well as in the combined pedigree (Fig 1). Pedigree A comprises the left side of the kindred, including the proband (III-13), her siblings (III-14 and III-15), their parents, and relatives of their mother (II-6). Pedigree B comprises the right half of the kindred, including the proband and her siblings, their parents, and relatives of their father (II-7).
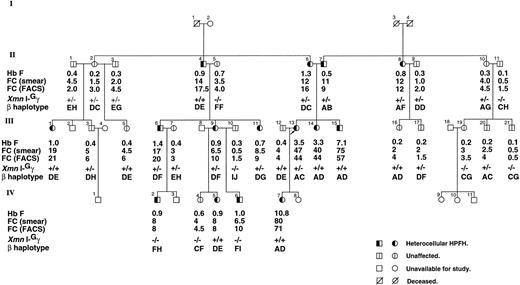
Pedigree of the English family with Hb F and F-cell levels by smear and FACS, β haplotype, and XmnI-Gγ site. β haplotype is constructed for 7 RFLPs, HindII-ε, HindIII-Gγ, HindIII-Aγ, HindII-Ψβ, HindII-3′Ψβ, AvaII-β, and BamHI-β, and denoted by A (−++−++++), B (+−−−−++), C (−++−+++), D (−+−+++−), E (−+−++++), F (+−−−−++), G (−++−++−), H (+−−−−+−), I (+−−−−−+), and J (−++++++). The γ gene triplication is found on the β chromosome associated with β haplotype A.

Pedigree of the English family with Hb F and F-cell levels by smear and FACS, β haplotype, and XmnI-Gγ site. β haplotype is constructed for 7 RFLPs, HindII-ε, HindIII-Gγ, HindIII-Aγ, HindII-Ψβ, HindII-3′Ψβ, AvaII-β, and BamHI-β, and denoted by A (−++−++++), B (+−−−−++), C (−++−+++), D (−+−+++−), E (−+−++++), F (+−−−−++), G (−++−++−), H (+−−−−+−), I (+−−−−−+), and J (−++++++). The γ gene triplication is found on the β chromosome associated with β haplotype A.
Multipoint analyses were undertaken with two sets of markers, D6S408-(0.023)-D6S407-(0.033)-D6S262 and D6S262-(0.066)-D6S976/D6S270-(0.018)-D6S292, using the program LINKMAP and FASTLINK20 21 in the combined pedigree and in both subpedigrees A and B independently. The figures in parentheses refer to the recombination fractions.
RESULTS
The English Pedigree
A 24-year-old white woman (III-13, Fig 1) was found to have an elevated level of Hb F following a Kleihauer test during her second pregnancy. Hb F remained elevated at 3.5% after the pregnancy and delivery of a normal female infant. The F-cell percentage was determined by immunofluorescence on peripheral blood smears and by flow cytometry to be 47% and 44%, respectively. The parents of the proband and her two younger siblings were studied, and it was found that both her brother (III-15) and sister (III-14) had clearly elevated levels of Hb F (7.1% and 3.3%, respectively). Hb F of the proband's mother (II-6) was also elevated at 1.3%, as was her F-cell percentage (12% and 16%, by peripheral blood smear and FACS, respectively). The father of the proband (II-7) had an apparently normal level of Hb F (0.5%), but the more sensitive immunostaining procedures yielded reproducibly elevated F-cell values (11% and 9% by peripheral blood smear and FACS). The Gγ:Aγ ratio of the propositus as determined by Triton-urea gel electrophoresis was approximately 0.5. The Gγ:Aγ ratio of the siblings of the propositus (III-14 and III-15) was determined by reverse-phase high-performance liquid chromatography. It was found that the common AγT variant was present, as well as AγI. The ratio of Gγ:AγT:AγI was 0.28:0.5:0.22 in the proband's sister (III-14) and 0.1:0.84:0.06 in her brother (III-15). The husband of the propositus (III-12) has normal levels of Hb F (0.4%) and F cells (4% by both methods). They have two daughters (IV-7 and IV-8), of whom only the older was available for study. At the age of 5 years, she has a marked elevation of Hb F (10.8%) in a heterocellular distribution (F cells, 80% and 71% by peripheral blood smear and FACS methods, respectively). The pedigree has been extended as far as possible, and relevant hematologic data are displayed in Fig 1 and Table 1. There is no known consanguinity in the family, and false paternity has been excluded.
Hematologic Data of Family Members
| Pedigree No. . | Sex/Age . | Hb (g/dL) . | Hb F (%) . | Hb A2 (%) . | F Cells (%) . | Xmn I-Gγ . | Aγ-bp Deletion . | |
|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | Smear . | FACS . | . | . |
| II-1 | M/64 | 15.8 | 0.4 | 2.3 | 4.5 | 2.0 | +/− | −/− |
| II-2 | F/62 | 14.4 | 0.2 | 2.2 | 1.5 | 3.0 | +/− | +/− |
| II-3 | M/70 | 15.4 | 0.3 | 2.2 | 2.0 | 4.5 | +/− | −/− |
| II-4 | M/58 | 14.8 | 0.9 | 3.0 | 14 | 17.5 | +/+ | −/− |
| II-5 | F/57 | 14.5 | 0.7 | 2.8 | 3.5 | 4.0 | −/− | −/− |
| II-6 | F/48 | 14.5 | 1.3 | 3.0 | 12 | 16 | +/− | +/− |
| II-7 | M/48 | 14.9 | 0.5 | 2.5 | 11 | 9 | +/− | −−*/− |
| II-8 | F/54 | 13.1 | 0.8 | 2.3 | 12 | 12 | +/− | −−*/− |
| II-9 | M/51 | 14.9 | 0.3 | 2.5 | 1.0 | 2.0 | +/− | −/− |
| II-10 | F/50 | 13.2 | 0.3 | 2.4 | 4.0 | 4.5 | +/− | −−*/+ |
| II-11 | M/53 | 17.0 | 0.1 | 2.5 | 0.5 | 1.5 | −/− | +/− |
| III-1 | F/40 | 14.5 | 1.0 | 1.9 | 19 | 21 | +/+ | −/− |
| III-3 | M/44 | 15.8 | 0.4 | 2.1 | 5.0 | 6.0 | +/− | −/− |
| III-5 | F/16 | 14.0 | 0.4 | 2.2 | 4.5 | 6.0 | +/+ | −/− |
| III-6 | M/36 | 16.3 | 1.4 | 2.5 | 17 | 20 | +/− | −/− |
| III-7 | F/34 | 14.0 | 0.4 | 2.8 | 3.0 | 3.0 | +/− | −/− |
| III-9 | F/33 | 12.3 | 0.9 | 3.0 | 6.5 | 10 | +/− | −/− |
| III-10 | M/32 | 16.3 | 0.3 | 2.2 | 0.5 | 1.5 | −/− | +/− |
| III-11 | F/33 | 11.1 | 0.7 | 2.5 | 8.5 | 9.0 | +/− | −/− |
| III-12 | M/36 | 15.2 | 0.4 | 2.8 | 4.0 | 4.0 | +/+ | −/− |
| III-13 | F/24 | 14.1 | 3.5 | 2.8 | 47 | 44 | +/− | −−*/+ |
| III-14 | F/22 | 14.1 | 3.3 | 2.9 | 40 | 44 | +/+ | −−*/− |
| III-15 | M/17 | 14.9 | 7.1 | 2.7 | 75 | 57 | +/+ | −−*/− |
| III-16 | F/18 | 13.8 | 0.2 | 2.5 | 2.0 | 4.0 | +/+ | −−*/− |
| III-17 | M/12 | 12.4 | 0.2 | 2.4 | 2.0 | 1.5 | +/− | −/− |
| III-19 | F/28 | 12.6 | 0.2 | 2.9 | 3.0 | 3.5 | −/− | +/+ |
| III-20 | M/27 | 17.1 | 0.2 | 2.4 | 2.5 | 4.0 | +/− | −−*/+ |
| III-21 | M/25 | 15.5 | 0.1 | 2.4 | 0.5 | 0.5 | −/− | +/+ |
| IV-2 | M/11 | 13.4 | 0.9 | 2.5 | 8.0 | 8.0 | −/− | −/− |
| IV-4 | F/16 | 14.3 | 0.6 | 3.0 | 4.0 | 4.5 | −/− | +/− |
| IV-5 | F/15 | 13.5 | 0.9 | 2.5 | 8.0 | 8.0 | +/+ | −/− |
| IV-6 | M/8 | 12.9 | 1.0 | 2.6 | 6.5 | 10 | −/− | −/− |
| IV-7 | F/5 | 12.6 | 10.8 | 2.9 | 80 | 71 | +/+ | −−*/− |
| Pedigree No. . | Sex/Age . | Hb (g/dL) . | Hb F (%) . | Hb A2 (%) . | F Cells (%) . | Xmn I-Gγ . | Aγ-bp Deletion . | |
|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | Smear . | FACS . | . | . |
| II-1 | M/64 | 15.8 | 0.4 | 2.3 | 4.5 | 2.0 | +/− | −/− |
| II-2 | F/62 | 14.4 | 0.2 | 2.2 | 1.5 | 3.0 | +/− | +/− |
| II-3 | M/70 | 15.4 | 0.3 | 2.2 | 2.0 | 4.5 | +/− | −/− |
| II-4 | M/58 | 14.8 | 0.9 | 3.0 | 14 | 17.5 | +/+ | −/− |
| II-5 | F/57 | 14.5 | 0.7 | 2.8 | 3.5 | 4.0 | −/− | −/− |
| II-6 | F/48 | 14.5 | 1.3 | 3.0 | 12 | 16 | +/− | +/− |
| II-7 | M/48 | 14.9 | 0.5 | 2.5 | 11 | 9 | +/− | −−*/− |
| II-8 | F/54 | 13.1 | 0.8 | 2.3 | 12 | 12 | +/− | −−*/− |
| II-9 | M/51 | 14.9 | 0.3 | 2.5 | 1.0 | 2.0 | +/− | −/− |
| II-10 | F/50 | 13.2 | 0.3 | 2.4 | 4.0 | 4.5 | +/− | −−*/+ |
| II-11 | M/53 | 17.0 | 0.1 | 2.5 | 0.5 | 1.5 | −/− | +/− |
| III-1 | F/40 | 14.5 | 1.0 | 1.9 | 19 | 21 | +/+ | −/− |
| III-3 | M/44 | 15.8 | 0.4 | 2.1 | 5.0 | 6.0 | +/− | −/− |
| III-5 | F/16 | 14.0 | 0.4 | 2.2 | 4.5 | 6.0 | +/+ | −/− |
| III-6 | M/36 | 16.3 | 1.4 | 2.5 | 17 | 20 | +/− | −/− |
| III-7 | F/34 | 14.0 | 0.4 | 2.8 | 3.0 | 3.0 | +/− | −/− |
| III-9 | F/33 | 12.3 | 0.9 | 3.0 | 6.5 | 10 | +/− | −/− |
| III-10 | M/32 | 16.3 | 0.3 | 2.2 | 0.5 | 1.5 | −/− | +/− |
| III-11 | F/33 | 11.1 | 0.7 | 2.5 | 8.5 | 9.0 | +/− | −/− |
| III-12 | M/36 | 15.2 | 0.4 | 2.8 | 4.0 | 4.0 | +/+ | −/− |
| III-13 | F/24 | 14.1 | 3.5 | 2.8 | 47 | 44 | +/− | −−*/+ |
| III-14 | F/22 | 14.1 | 3.3 | 2.9 | 40 | 44 | +/+ | −−*/− |
| III-15 | M/17 | 14.9 | 7.1 | 2.7 | 75 | 57 | +/+ | −−*/− |
| III-16 | F/18 | 13.8 | 0.2 | 2.5 | 2.0 | 4.0 | +/+ | −−*/− |
| III-17 | M/12 | 12.4 | 0.2 | 2.4 | 2.0 | 1.5 | +/− | −/− |
| III-19 | F/28 | 12.6 | 0.2 | 2.9 | 3.0 | 3.5 | −/− | +/+ |
| III-20 | M/27 | 17.1 | 0.2 | 2.4 | 2.5 | 4.0 | +/− | −−*/+ |
| III-21 | M/25 | 15.5 | 0.1 | 2.4 | 0.5 | 0.5 | −/− | +/+ |
| IV-2 | M/11 | 13.4 | 0.9 | 2.5 | 8.0 | 8.0 | −/− | −/− |
| IV-4 | F/16 | 14.3 | 0.6 | 3.0 | 4.0 | 4.5 | −/− | +/− |
| IV-5 | F/15 | 13.5 | 0.9 | 2.5 | 8.0 | 8.0 | +/+ | −/− |
| IV-6 | M/8 | 12.9 | 1.0 | 2.6 | 6.5 | 10 | −/− | −/− |
| IV-7 | F/5 | 12.6 | 10.8 | 2.9 | 80 | 71 | +/+ | −−*/− |
Presence of the 4-bp deletion at positions −221 to −224 of the Aγ gene is noted by +.
Denotes β cluster allele carrying the γ gene triplication (γγγ).
A total of 33 individuals all older than 5 years from three generations have been studied. No members of the pedigree were anemic. In each case, red blood cell indices were normochromic normocytic, and Hb A2 levels were within the normal range. There was no evidence of β- or α-thalassemia. Further individuals on both sides of the extended family have elevated F-cell percentages, although none are as pronounced as those present in the proband, her two siblings, or her daughter.
Gene mapping showed the β-globin gene complex to be intact with no evidence of a deletion within the β cluster. A γ gene triplication on one allele was revealed from DNA analysis and confirmed by BglII-γ restriction mapping as previously described.22 Nine members of the extended family were heterozygous for the γ gene triplication (γγγ) as shown in Fig 1 (β haplotype A) and Table 1. Sequence analysis of the Gγ and Aγ promoter regions of all five members of the proband's nuclear family did not show any difference from published normal sequences. Two sequence variations were found: the T-C polymorphism at position −158 of the Gγ gene (detected by XmnI restriction analysis)15,16 and a 4-bp deletion at positions −221 to −224 of the Aγ gene (detected by Fnu4HI restriction analysis)23-26 (Table 1).
Considering all the information available, there is evidence of heterocellular HPFH segregating in this family in the absence of any hemoglobinopathy, although the Hb F value of 7.1% in III-15 and 10.8% in IV-7 is higher than the level normally associated with this form of HPFH. Karyotype analysis was performed on the members of the immediate family of the propositus. No abnormality was found.
Inheritance Pattern of Heterocellular HPFH
Hb F levels in the 33 family members range from 0.1% to 10.8%, which corresponds to F-cell values of 0.5% to 80%. Four individuals have Hb F levels more than 3.0% and F-cell values more than 40% (the propositus, her two siblings, and her daughter). These individuals have been studied on three separate occasions with consistent results. By contrast, the other affected individuals have Hb F levels of less than 1.5% and F-cell levels ranging from 5% to 21%. There are 15 individuals with F-cell values greater than 8% (according to FACS analysis). In these cases, there is a strong likelihood that a HPFH determinant exists. However, four individuals are reproducibly in the F-cell range of 4% to 6% (II-10, III-3, III-5, and IV-4), which emphasizes the difficulty in drawing an arbitrary line between individuals unaffected by a HPFH determinant (but at the upper end of the “normal” distribution) and those who are affected by the HPFH determinant but show only a modest elevation of F cells. These individuals have also been studied on two separate occasions, and consistent results were obtained.
Statistical analysis using the least-squares method in a previous survey8 of 300 healthy adults has suggested that individuals with at least 4.4% F cells may be considered affected with HPFH. However, in view of the difficulty in drawing an arbitrary cutoff point, for linkage purposes in this study, individuals with F cells more than 4% and less than 8% were classified as unaffected but assigned to liability class 2.
Incomplete penetrance.Individual II-2 is a maternal aunt of the propositus and has a Hb F level of 0.2% with F-cell values of 1.5% and 3.0% by the peripheral blood smear and FACS methods, respectively. Her two siblings (II-4 and II-6) have F-cell values of 17.5% and 16% (by FACS), respectively. II-2 has three offspring (by two fathers: II-1 and II-3) who were available for study. Of the three children, one is clearly affected (III-1, F cells 21% by FACS) and the other two have F-cell percentages between 4.5% and 6%. The phenotype of the clearly affected daughter (III-1) is very similar to that of her maternal uncle (II-4) and aunt (II-6), and it therefore seems likely that individual II-2 has the HPFH genotype and has passed it to her daughter without expressing the phenotype herself. The phenotypes of the individuals concerned were reproducible. There are no other examples of incomplete penetrance in this pedigree.
There is father-to-son transmission of HPFH.There are three occasions in this pedigree in which an affected male has had children who were available for study (II-4, II-7, and III-6). In each case, there has been an affected son (II-4 to III-6, III-6 to IV-2, and II-7 to III-15). II-7 and his son III-15 should probably not be considered as evidence of father-to-son transmission, as the HPFH determinant could have been passed to III-15 by his affected mother (II-6). However, the case of II-4 (F cells 17.5% by FACS) transmitting the trait to his son III-6 (F cells 20% by FACS) is good evidence against X-linkage.
The phenotype segregates independently of the β-globin complex but the Xmn I-Gγ polymorphism may be involved in expression of the trait.The β haplotypes, Aγ4-bp deletion polymorphism, and the γ gene triplication served as informative markers for segregation of the β-globin complex, and evidence for independent segregation of the HPFH determinant was provided in several instances.
The propositus (III-13), her two siblings (III-14 and III-15), her daughter (IV-7), her father (II-7), and her paternal aunt (II-8) are all clearly affected, and all are carrying the γ gene triplication associated with haplotype A (Fig 1). However, individuals III-16, II-10, and III-20 are also carrying the same β allele and are unaffected. Thus, there is good evidence that the phenotype is not linked to the γ gene triplication haplotype. It has been shown previously that the γ gene triplication arrangement is not associated with increased Hb F levels.22 27
In the other half of the pedigree, inspection of Fig 1 shows that if the HPFH determinant behaves as an allele of the β-globin complex, it must be carried on a chromosome associated with either the C or D haplotype. Of the affected family members, one (III-13) has the C haplotype, eight have the D haplotype, and one has both (II-6). In three instances (III-13 to IV-7, III-9 to IV-6, and III-6 to IV-2), transmission of the HPFH phenotype has occurred without the C or D haplotype.
XmnI-Gγ polymorphism has been shown to influence the level of F cells in normal individuals.6 In this pedigree, while 13 of 15 affected individuals are either XmnI-Gγ +/+ or +/−, the other two affected individuals (IV-2 and IV-6) are XmnI-Gγ −/−, thereby providing evidence that the phenotype in this family is not simply (or completely) related to the presence of the XmnI-Gγ site. It is likely that the extent to which the HPFH phenotype is expressed is dependent on the XmnI-Gγ genotype (as we have previously demonstrated in a large Asian-Indian pedigree4 ).
Linkage Analysis With Polymorphic Markers on 6q and the β-Globin Complex
Two-point Lod scores were determined between the phenotype and the polymorphic markers in the 6q22-q23 region and in the β-globin complex (Table 2). Since several affected individuals in pedigree A were homozygous for the marker D6S976 that is tightly linked to marker D6S270, haplotypes were generated for these two markers and analysis was performed using the D6S976/D6S270 haplotype to increase the informativeness for linkage. Individuals with F-cell values of at least 8.0% were considered to be affected and assigned to liability class 1, except for individuals III-13, III-14, and III-15, who have very high F-cell levels and were assumed to have inherited the HPFH determinants from both parents. Individuals III-13, III-14, and III-15 were assigned to liability class 3. Individuals II-10, III-3, III-5, and IV-4 have F-cell values of 4.5% to 6.0% as determined by FACS. These individuals were assigned to unaffected status (liability class 2). The phenotype assigned to the individuals is indicated in Fig 1. No adjustment has been made for sex, or the XmnI-Gγ status of individuals in this preliminary study.
Two-Point Lod Score for the Microsatellite in the β-Globin Gene Cluster and the Chromosome 6q Markers in the Combined Pedigree and the Two Halves, Pedigrees A and B
| Marker . | Lod Score at Recombination Fraction (θ) of . | ||||||
|---|---|---|---|---|---|---|---|
| . | .00 . | .01 . | .05 . | .10 . | .20 . | .30 . | .40 . |
| β cluster | |||||||
| Combined | −infini | −2.17 | −0.88 | −0.42 | −0.11 | −0.03 | −0.02 |
| Pedigree A | −infini | −2.51 | −1.18 | −0.67 | −0.27 | −0.11 | −0.04 |
| Pedigree B | 0.35 | 0.34 | 0.30 | 0.25 | 0.16 | 0.08 | 0.02 |
| D6S408 | |||||||
| Combined | −4.24 | −2.40 | −0.76 | −0.15 | 0.21 | 0.22 | 0.12 |
| Pedigree A | −3.05 | −1.08 | −0.06 | 0.27 | 0.38 | 0.28 | 0.14 |
| Pedigree B | −3.34 | −1.94 | −0.84 | −0.38 | −0.04 | 0.04 | 0.02 |
| D6S407 | |||||||
| Combined | −infini | −4.77 | −2.93 | −1.94 | −0.83 | −0.28 | −0.05 |
| Pedigree A | −infini | −3.62 | −2.10 | −1.37 | −0.56 | −0.17 | −0.02 |
| Pedigree B | −3.06 | −1.92 | −1.01 | −0.55 | −0.15 | −0.02 | 0.01 |
| D6S262 | |||||||
| Combined | −infini | −4.41 | −1.93 | −0.89 | −0.06 | 0.15 | 0.11 |
| Pedigree A | −infini | −3.01 | −1.21 | −0.43 | 0.13 | 0.22 | 0.12 |
| Pedigree B | −5.30 | −2.17 | −0.90 | −0.42 | −0.07 | 0.02 | 0.02 |
| D6S435 | |||||||
| Combined | −infini | −5.31 | −2.53 | −1.33 | −0.37 | −0.07 | −0.00 |
| Pedigree A | −infini | −3.23 | −1.34 | −0.57 | −0.03 | 0.06 | 0.03 |
| Pedigree B | −7.48 | −2.85 | −1.37 | −0.74 | −0.22 | −0.04 | 0.00 |
| D6S457 | |||||||
| Combined | −5.89 | −2.33 | −1.39 | −0.82 | −0.18 | 0.05 | 0.07 |
| Pedigree A | −0.25 | −0.26 | −0.20 | −0.05 | 0.16 | 0.19 | 0.10 |
| Pedigree B | −7.44 | −2.85 | −1.37 | −0.74 | −0.22 | −0.04 | 0.00 |
| D6S413 | |||||||
| Combined | −infini | −3.06 | −2.12 | −1.49 | −0.71 | −0.28 | −0.07 |
| Pedigree A | −infini | −2.83 | −1.94 | −1.36 | −0.64 | −0.25 | −0.06 |
| Pedigree B | −2.63 | −1.58 | −0.89 | −0.58 | −0.26 | −0.11 | −0.03 |
| D6S472 | |||||||
| Combined | −infini | −3.35 | −1.79 | −1.11 | −0.44 | −0.12 | 0.00 |
| Pedigree A | −infini | −2.24 | −1.37 | −0.85 | −0.32 | −0.08 | 0.01 |
| Pedigree B | −2.70 | −1.38 | −0.72 | −0.43 | −0.17 | −0.06 | −0.01 |
| D6S975 | |||||||
| Combined | −infini | −4.29 | −2.01 | −1.02 | −0.23 | 0.02 | 0.03 |
| Pedigree A | −infini | −2.22 | −0.82 | −0.26 | 0.12 | 0.15 | 0.06 |
| Pedigree B | −5.00 | −1.49 | −0.66 | −0.30 | −0.03 | 0.04 | 0.02 |
| D6S976/D6S270 | |||||||
| Combined | −5.69 | −1.35 | −0.00 | 0.49 | 0.72 | 0.58 | 0.30 |
| Pedigree A | −0.05 | 0.73 | 1.19 | 1.25 | 1.06 | 0.72 | 0.33 |
| Pedigree B | −5.04 | −1.49 | −0.66 | −0.30 | −0.03 | 0.04 | 0.02 |
| D6S292 | |||||||
| Combined | −4.14 | −2.46 | −1.38 | −0.76 | −0.12 | 0.12 | 0.13 |
| Pedigree A | −1.30 | −1.21 | −0.86 | −0.47 | 0.02 | 0.17 | 0.14 |
| Pedigree B | −2.88 | −1.54 | −0.82 | −0.50 | −0.20 | −0.07 | −0.01 |
| Marker . | Lod Score at Recombination Fraction (θ) of . | ||||||
|---|---|---|---|---|---|---|---|
| . | .00 . | .01 . | .05 . | .10 . | .20 . | .30 . | .40 . |
| β cluster | |||||||
| Combined | −infini | −2.17 | −0.88 | −0.42 | −0.11 | −0.03 | −0.02 |
| Pedigree A | −infini | −2.51 | −1.18 | −0.67 | −0.27 | −0.11 | −0.04 |
| Pedigree B | 0.35 | 0.34 | 0.30 | 0.25 | 0.16 | 0.08 | 0.02 |
| D6S408 | |||||||
| Combined | −4.24 | −2.40 | −0.76 | −0.15 | 0.21 | 0.22 | 0.12 |
| Pedigree A | −3.05 | −1.08 | −0.06 | 0.27 | 0.38 | 0.28 | 0.14 |
| Pedigree B | −3.34 | −1.94 | −0.84 | −0.38 | −0.04 | 0.04 | 0.02 |
| D6S407 | |||||||
| Combined | −infini | −4.77 | −2.93 | −1.94 | −0.83 | −0.28 | −0.05 |
| Pedigree A | −infini | −3.62 | −2.10 | −1.37 | −0.56 | −0.17 | −0.02 |
| Pedigree B | −3.06 | −1.92 | −1.01 | −0.55 | −0.15 | −0.02 | 0.01 |
| D6S262 | |||||||
| Combined | −infini | −4.41 | −1.93 | −0.89 | −0.06 | 0.15 | 0.11 |
| Pedigree A | −infini | −3.01 | −1.21 | −0.43 | 0.13 | 0.22 | 0.12 |
| Pedigree B | −5.30 | −2.17 | −0.90 | −0.42 | −0.07 | 0.02 | 0.02 |
| D6S435 | |||||||
| Combined | −infini | −5.31 | −2.53 | −1.33 | −0.37 | −0.07 | −0.00 |
| Pedigree A | −infini | −3.23 | −1.34 | −0.57 | −0.03 | 0.06 | 0.03 |
| Pedigree B | −7.48 | −2.85 | −1.37 | −0.74 | −0.22 | −0.04 | 0.00 |
| D6S457 | |||||||
| Combined | −5.89 | −2.33 | −1.39 | −0.82 | −0.18 | 0.05 | 0.07 |
| Pedigree A | −0.25 | −0.26 | −0.20 | −0.05 | 0.16 | 0.19 | 0.10 |
| Pedigree B | −7.44 | −2.85 | −1.37 | −0.74 | −0.22 | −0.04 | 0.00 |
| D6S413 | |||||||
| Combined | −infini | −3.06 | −2.12 | −1.49 | −0.71 | −0.28 | −0.07 |
| Pedigree A | −infini | −2.83 | −1.94 | −1.36 | −0.64 | −0.25 | −0.06 |
| Pedigree B | −2.63 | −1.58 | −0.89 | −0.58 | −0.26 | −0.11 | −0.03 |
| D6S472 | |||||||
| Combined | −infini | −3.35 | −1.79 | −1.11 | −0.44 | −0.12 | 0.00 |
| Pedigree A | −infini | −2.24 | −1.37 | −0.85 | −0.32 | −0.08 | 0.01 |
| Pedigree B | −2.70 | −1.38 | −0.72 | −0.43 | −0.17 | −0.06 | −0.01 |
| D6S975 | |||||||
| Combined | −infini | −4.29 | −2.01 | −1.02 | −0.23 | 0.02 | 0.03 |
| Pedigree A | −infini | −2.22 | −0.82 | −0.26 | 0.12 | 0.15 | 0.06 |
| Pedigree B | −5.00 | −1.49 | −0.66 | −0.30 | −0.03 | 0.04 | 0.02 |
| D6S976/D6S270 | |||||||
| Combined | −5.69 | −1.35 | −0.00 | 0.49 | 0.72 | 0.58 | 0.30 |
| Pedigree A | −0.05 | 0.73 | 1.19 | 1.25 | 1.06 | 0.72 | 0.33 |
| Pedigree B | −5.04 | −1.49 | −0.66 | −0.30 | −0.03 | 0.04 | 0.02 |
| D6S292 | |||||||
| Combined | −4.14 | −2.46 | −1.38 | −0.76 | −0.12 | 0.12 | 0.13 |
| Pedigree A | −1.30 | −1.21 | −0.86 | −0.47 | 0.02 | 0.17 | 0.14 |
| Pedigree B | −2.88 | −1.54 | −0.82 | −0.50 | −0.20 | −0.07 | −0.01 |
Linkage with the β-globin cluster is excluded in the combined pedigree and pedigree A (Lod score < −2) at θ less than .01 (Table 2). The data suggest that the genetic determinant causing HPFH is located outside the β-globin gene complex and support previous investigations of the pedigree in which no mutation associated with HPFH has been found in the β-globin cluster.
The hypothesis of linkage of the HPFH phenotype to the 6q22.3-q23.2 region was similarly tested. Multipoint analysis provides no evidence for linkage to the 6q candidate region.
Multilocus linkage analysis was undertaken in pedigrees A and B both independently and combined (Fig 2). Calculations were confined to four-point analysis using the HPFH locus and two sets of markers in the 6q region — D6S408, D6S407, and D6S262 and D6S262, D6S976/D6S270, and D6S292. The results confirm that there is no evidence for the location of the HPFH gene within the 6q region spanned by D6S408 and D6S292.

Multipoint Lod scores for the location of the HPFH locus in relation to 2 sets of markers in the 6q22.3-q23.2 region: (A) D6S408, D6S407, D6S262 and (B) D6S262, D6S976/D6S270, D6S292. Results are plotted as a function of distance from D6S408 (A) or D6S262 (B). Analyses were undertaken in the combined pedigree and independently in each half, pedigree A and pedigree B. Recombination fractions were converted to cM using the Haldane map function.

Multipoint Lod scores for the location of the HPFH locus in relation to 2 sets of markers in the 6q22.3-q23.2 region: (A) D6S408, D6S407, D6S262 and (B) D6S262, D6S976/D6S270, D6S292. Results are plotted as a function of distance from D6S408 (A) or D6S262 (B). Analyses were undertaken in the combined pedigree and independently in each half, pedigree A and pedigree B. Recombination fractions were converted to cM using the Haldane map function.
DISCUSSION
The mode of inheritance of unlinked HPFH has been variously described as autosomal dominant, autosomal codominant, X-linked, and polygenic.28,29 An extended English family with heterocellular HPFH has been studied in an effort to gain insight into the phenotype and inheritance pattern of this condition. The family presented is larger than other published pedigrees with heterocellular HPFH segregating in the absence of any hemoglobinopathy despite which a complex situation has emerged. In this English pedigree, X-linkage is unlikely considering that father-to-son transmission has occurred. The pattern of transmission from II-4 to III-6 to IV-2 (in each case, a male with an unaffected partner) is suggestive of autosomal dominant inheritance. However, there are complexities that remain unexplained, such as variability in the extent of Hb F and F-cell elevation (variable expressivity) and incomplete penetrance. The tremendous variability in the elevation of Hb F and F cells is not unique to this family, but is also previously observed in other families with HPFH.4 The HPFH is not linked to the β-globin gene complex, although the XmnI-Gγ site appears to modify the phenotype; the most markedly affected individuals have at least one XmnI-Gγ site. However, the association between affected status and the presence of this polymorphism is not absolute. Autosomal codominant transmission is a strong possibility and would explain the more pronounced elevations of Hb F and F cells observed in individuals III-13, III-14, and III-15, who would be considered homozygotes or compound heterozygotes for two HPFH determinants. Their parents (II-6 and II-7) are both affected with modest elevations of F cells. It is not possible in this family to determine if HPFH determinants are allelic or nonallelic.
The limited published data regarding heterocellular HPFH in whites in the absence of hemoglobinopathies support the results obtained in this family. The original study by Marti30 and the population survey by Zago et al7 established that the Hb F (and F cell) levels of normal individuals are genetically determined. They traced the parents and siblings of probands with Hb F levels at the upper limits of the population range. In the majority of cases, one of the parents had a similarly increased Hb F (or F cell) level. In both studies, there was at least one example in which neither parent displayed elevated levels of Hb F. Assuming true paternity, this may represent incomplete penetrance of the trait, as found on one occasion in the English pedigree.
The hypothesis that heterocellular HPFH in this English pedigree is determined by the same gene(s) in 6q responsible for heterocellular HPFH in the large Asian-Indian pedigree11 has been examined. The results obtained strongly suggest that this region is an unlikely candidate for a locus associated with the HPFH phenotype in the English pedigree. The results of linkage analysis using the β-globin cluster microsatellite marker17 confirm the absence of linkage between the β cluster and the HPFH phenotype.
It was hoped that the clear-cut phenotype of the proband in this pedigree would be present in other members of the extended family. However, the situation as described has established that this form of HPFH is indeed complex and genetically heterogeneous and that, apart from the β-globin locus and the 6q gene, there must be at least one other autosomal locus that controls Hb F levels in adults. The underlying genetic heterogeneity underscores the importance of analyzing data from one large pedigree, since interpretation of linkage analyses involving several different small families is difficult and could be misleading.
ACKNOWLEDGMENT
We thank Liz Rose and Milly Graver for preparation of the manuscript, Professor Peter Beverley for permission to use the anti–γ-globin chain antibody, and Professor Sir D.J. Weatherall for encouragement and support. Linkage analyses were undertaken using programs provided by the UK Human Genome Mapping Project Resource Centre (funded by the MRC).
Supported by a Nuffield Dominion Fellowship, a European Community (EC) Senior Fellowship, a Wellcome European Fellowship, and a Wellcome Senior Clinical Fellowship.
Address reprint requests to S.L. Thein, MD, MRC Molecular Hematology Unit, Institute of Molecular Medicine, John Radcliffe Hospital, Headington, Oxford OX3 9DU, UK.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal