Abstract
Antisense oligodeoxyribonucleotides (ODNs) are now being extensively investigated in an attempt to achieve cell growth suppression through specific targeting of genes related to cell proliferation, despite increasing evidence of non-antisense cytotoxic effects. In the context of anti-BCR/ABL antisense strategies in chronic myeloid leukemia, we have re-examined the antiproliferative effect of phosphodiester and phosphorothioate ODNs on the leukemic cell line BV173 and on CD34+ bone marrow cells in liquid culture. The 3′ sequences of the ODNs determine their effect. At concentrations of 10 μmol/L (for phosphorothioate ODNs) or 25 μmol/L (for phosphodiester ODNs), all the tested ODNs exert an antiproliferative activity, except those that contain a cytosine residue at either their two most terminal 3′ positions. We show that this antiproliferative effect is due to the toxicity of the d-NMPs (5′ monophosphate deoxyribonucleosides), the enzymatic hydrolysis products of the ODNs in culture medium. The toxicity of the d-NMPs on hematologic cells depends on their nature (d-CMP [2′deoxycytidine 5′-monophosphate] is not cytotoxic), on their concentration (d-GMP [2′-deoxyguanosine 5′-monophosphate], TMP [thymidine 5′-monophosphate], and d-AMP [2′-deoxyadenosine 5′-monophosphate] are cytotoxic at concentrations between 5 and 10 μmol/L), and on the coincident presence of other d-NMPs in the culture medium (d-CMP neutralizes the toxicity of d-AMP, d-GMP, or TMP). The antiproliferative activity of ODNs is thus restricted to conditions where the 3′ hydrolysis process by exonucleases generates significant amounts of d-NMPs with a low proportion of d-CMP. Our results reveal a novel example of a nonantisense effect of ODNs, which should be taken into account when performing any experiment using assumed antisense ODNs.
AN INCREASING number of publications are now pointing out the difficulty in interpreting the oligodeoxyribonucleotides (ODN)-mediated antisense studies.1-6 Many biological effects of ODNs cannot be attributed to sequence-specific inhibition of genetic expression. The aptameric properties of ODNs (sequence-specific interactions between ODNs and molecules other than nucleic acids) may be responsible for some biological effects.7-9 The non-sequence-specific binding of ODNs, which are polyanions, to proteins may disturb important physiological processes.10,11 The degradation products of the ODNs may also induce effects.2
We recently analyzed12 the effect of antisense phosphodiester ODNs,13-33 complementary to the junctional region of the BCR/ABL mRNA,34-39 on the proliferation of leukemic cells and cell lines. Using numerous control ODNs (sense, mismatched, scrambled, and random control sequences), we have shown that the ODN induced antiproliferative result is a sequence-specific but non-antisense effect. We have further observed that this effect is not leukemia-specific. A “TAT” triplet at the 3′ end of the ODN sequences was found to be sufficient to induce an antiproliferative effect, regardless of the remaining bases.
In this study, we analyzed the mechanism responsible for this sequence-specific effect of phosphodiester ODNs on the leukemic BV173 cell line and on CD34+ bone marrow cells in tissue culture. We also examined whether, under similar experimental conditions, the reported antiproliferative effect of anti-BCR/ABL phosphorothioate ODNs21 can also be assigned to their terminal 3′ sequences.
MATERIALS AND METHODS
Chemicals.d-AMP (2′-deoxyadenosine 5′-monophosphate), d-CMP (2′-deoxycytidine 5′-monophosphate), d-GMP (2′-deoxyguanosine 5′-monophosphate) and TMP (thymidine 5′-monophosphate), all for laboratory use (purity 98%), were obtained from Sigma Chemical Co (St Louis, MO). Sodium chloride (analysis grade), tris-hydroxymethylaminomethane (for molecular biology) and acetonitrile (for chromatography) were purchased from Merck (Darmstadt, Germany). Ammonium hydroxide (28% to 30% wt/vol, analysis grade) was purchased from Acros Organics (Geel, Belgium). Absolute ethyl alcohol (analysis grade) was obtained from UCB (Leuven, Belgium). RPMI 1640 culture medium and fetal calf serum (FCS), both for cell culture, were from GIBCO-BRL (Scotland, UK). Normal human serum (NHS) was obtained from the blood of healthy donors.
ODNs.Phosphodiester, phosphorothioate, and chimeric phosphodiester/phosphorothioate ODNs were synthesized on an automated DNA synthesizer (Cyclone Plus DNA synthesizer or Expedite 8909 DNA/RNA synthesizer, MilliGen/Biosearch, Millipore, Burlington, MA) using the β-cyanoethyl phosphoramidite chemistry, according to the manufacturer's protocols. After synthesis, the ODNs were cleaved from their solid support and deblocked in 30% ammonium hydroxide at 55°C for 16 hours. The phosphodiester and chimeric ODNs were purified by anion exchange chromatography40 (column MONO Q HR 5/5; Pharmacia, Uppsala, Sweden) or Protein-Pack Q 8HR, Waters, Milford, MA) and desalted (NAP columns; Pharmacia Biotech, Uppsala, Sweden). Phosphodiester and chimeric phosphodiester/phosphorothioate ODNs were evaluated from analytical high-performance liquid chromatography (HPLC) (see conditions below) to be >95% full-length material. Phosphorothioate ODNs were purified on reverse phase column (column pep RPC, Pharmacia) via DMT-ON (5′-O-dimethoxytrityl-on) procedure, according to the manufacturer's protocol. Fully deprotected phosphorothioate ODNs were then precipitated twice from a 0.5 mol/L NaCl solution with 2.5 volumes ethanol, and then desalted (NAP columns; Pharmacia Biotech). Purified phosphorothioate ODNs were greater than 95% full-length material as determined by polyacrylamide gel electrophoresis. Some of the ODNs were kindly supplied by GENSET (Paris, France) or EUROGENTEC (Liège, Belgium) and were purified by the procedures described above. The ODNs were stored in water at −20°C. Different batches of ODNs from different origins were used to avoid effects due to a particular synthesis and/or purification.
HPLC analysis.HPLC analyses were performed on a Waters instrument, equipped with a 625 PEEK solvent delivery system, a 717 autosampler, a 486 UV detector set to 260 nm, and a Digital Celebris FP 590 computer (Maynard, MA). The Millenium 2.1 software (Waters) was used to drive the instruments and to analyze the chromatograms. The chromatography of the ODN samples was performed on an anion exchange column (NucleoPac PA-100 and NucleoPac PA-100 Guard columns; Dionex, Sunnyvale, CA) using a gradient of NaCl (10 mmol/L NaCl/min) in 25 mmol/L Tris-HCl, pH 8.0, with a flow of 1.5 mL/min. The duration of the gradient (typically 20 minutes) and the salt concentration at the start of elution (typically 300 mmol/L NaCl) were adapted to the test samples.
Kinetics of ODN degradation in cell culture media.Phosphodiester or chimeric phosphodiester/phosphorothioate ODNs were incubated at 37°C at a concentration of 25 μmol/L in RPMI 1640 medium containing 2.5% NHS or 10% FCS. Samples were collected at various time intervals during incubation. After addition of an internal standard (a 23-mer phosphodiester ODN), the samples were stored immediately at −20°C. Before HPLC analysis (see conditions above), the crude samples were filtrated on low-binding regenerated cellulose Ultrafree-MC units (10,000 NMWL Filter Unit; Millipore). The area of each chromatographic signal (measured at 260 nm) was normalized to the area of the internal standard.
Cell line.The human leukemia cell line BV17341 was maintained by serial passages (twice weekly) in RPMI 1640 medium supplemented with 10% FCS, 2 mmol/L L-glutamine (GIBCO-BRL), 100 μg/mL streptomycin (Diamant, Compiègne, France) and 100 IU/mL penicillin (Continental Pharma, Brussels, Belgium). Cells were incubated at 37°C in 100-mL tissue culture flasks in a humidified atmosphere with 5% CO2. The cell line was controlled by reverse transcription-polymerase chain reaction for the expression of the normal ABL and BCR genes and of the b2a2 BCR/ABL fusion gene, as previously described.42
ODN and d-NMP treatment of cell line.Cells in their exponential phase of growth were washed with RPMI 1640 medium, and resuspended in RPMI 1640 containing 2.5% NHS, at a concentration of 105 cells/mL. The cell suspension was distributed into 2-mL aliquots. The lyophilized ODNs (20 nmol for phosphorothioate ODNs and 50 nmol for phosphodiester) were dissolved in cell suspensions, or d-NMPs (20 to 100 μL of 2.5 to 0.1 mmol/L d-NMP solutions in RPMI 1640) were added to the cell suspensions at once, at time 0 of each experiment. No further addition of d-NMPs was made. Viable cell number was determined every 24 hours on 100-μL aliquots using a trypan blue dye exclusion test in a Bürker hemocytometer. The control consisted of cells undergoing the same procedure, without addition of ODNs or d-NMPs.
Separation of normal CD34+ cells.Bone marrow cells from 3 healthy donors were separated by density gradient centrifuging at 600g for 20 minutes using a lymphocyte separation medium (International Medical Belgium, Brussels, Belgium). The samples were enriched in CD34+ cells using a Ceprate LC34 column (CellPro, Bothell, WA), according to the manufacturer's instructions. The CD34+ fractions were resuspended at a cell concentration of 105 cell/mL in RPMI 1640 containing 2.5 % NHS, 2 mmol/L L-glutamine, 50 ng/mL rhu-IL-3 (Sandoz, Basel, Switzerland), 20 ng/mL rhu-SCF (Immunex, Seattle, WA) and 100 ng/mL rhu-IL-6 (Sandoz). The cell suspensions were then distributed into 2-mL aliquots which were used for the ODN and d-NMP incubation experiments after 24 hours of growth. ODNs or d-NMPs were added to the cell suspensions at once. No further addition of d-NMPs was made.
ODN and d-NMP treatment of marrow cells.The lyophilized ODNs (20 nmol for phosphorothioate ODNs and 50 nmol for phosphodiester ODNs) were dissolved in the bone marrow cell suspensions. d-NMPs (20 to 100 μL from 2.5 to 0.1 mmol/L d-NMP solutions in RPMI 1640) were added to the marrow cell suspensions. The cells were counted every 24 hours for 4 days using the trypan blue dye exclusion test. The control consisted of cells undergoing the same procedure, without addition of ODNs or d-NMPs.
RESULTS
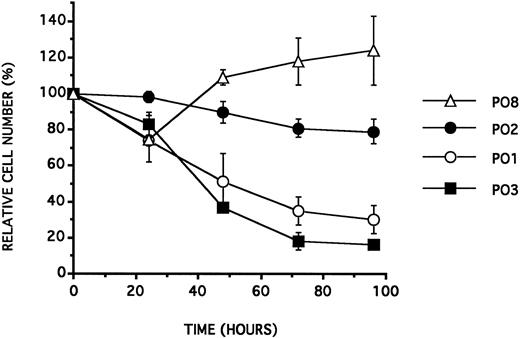
Sequence specificity and antiproliferative effect on the BV173 cell line.We incubated the BV173 cell line with the 18 mer phosphodiester ODNs PO1 to PO12 presented in Table 1. The ODN PO1 was synthesized with a sequence complementary to the b2a2 junction of the BCR/ABL mRNA. The ODNs PO2 to PO9 were designed with their sequence identical to PO1 except for a one base change at one of the three last 3′ positions. The ODNs PO10 and PO11 were constructed with the same base composition but not the same sequence as PO1. PO12 had a sequence complementary to PO1. All the experiments were performed at an ODN concentration of 25 μmol/L and in a culture medium supplemented with 2.5% NHS. As previously reported,12 under such conditions, the ODNs containing a TAT at their 3′ end, such as PO1, generated an antiproliferative effect, after 72 hours of incubation with the cells. As reported in Table 2 and in Fig 1, all the tested ODNs showed the same antiproliferative effect, except those containing a cytosine residue at either their two most terminal positions.
ODNs Used in This Study
| ODN . | Sequence (5′-3′) . | Half-Life (Hour) . | |
|---|---|---|---|
| . | . | 2.5% NHS . | 10% FCS . |
| PO1 | GAAGGGCTTCTTCCTTAT | 22 ± 3 | <1 |
| PO2 | GAAGGGCTTCTTCCTTCT | 38 ± 4 | <1 |
| PO3 | GAAGGGCTTCTTCCTTGT | 19 ± 1 | |
| PO4 | GAAGGGCTTCTTCCTTTT | 17 ± 1 | |
| PO5 | GAAGGGCTTCTTCCTTAA | ||
| PO6 | GAAGGGCTTCTTCCTCAT | 24 ± 1 | |
| PO7 | GAAGGGCTTCTTCCTGAT | 29 ± 2 | |
| PO8 | GAAGGGCTTCTTCCTTAC | 32 ± 1 | |
| PO9 | GAAGGGCTTCTTCCTTAG | 47 ± 2 | |
| PO10 | AGTCTTGCATCGTCTTGA | ||
| PO11 | ATTCTGTGAAGTCCTTGC | ||
| PO12 | ATAAGGAAGAAGCCCTTC | 37 ± 4 | |
| PO13 | TTTTTTTTTTTTTTTTTT | 12 ± 1 | |
| PO14 | CCCCCCCCCCCCCCCCCC | >72 | |
| PO/PS1 | GAAGGGCTTCTTCCTTA(Ps)T | >100 | 15 ± 1 |
| PO/PS2 | GAAGGGCTTCTTCCTT(pS)AT | 23 ± 4 | <1 |
| PO/PS3 | GAAGGGCTTCTTCCT(pS)TAT | 25 ± 2 | <1 |
| PS1 | CGCTGAAGGGCTTCTTCCTTATTGAT | ||
| PS2 | ATCAATAAGGAAGAAGCCCTTCAGCG | ||
| PS3 | CGCTGAAGGGCTTTTGAACTCTGCTT | ||
| PS4 | AAGCAGAGTTCAAAAGCCCTTCAGCG | ||
| PS5 | CGCTGAAGGGCTTCTTCCTTATTGCG | ||
| PS6 | ATCAATAAGGAAGAAGCCCTTCAGAT | ||
| ODN . | Sequence (5′-3′) . | Half-Life (Hour) . | |
|---|---|---|---|
| . | . | 2.5% NHS . | 10% FCS . |
| PO1 | GAAGGGCTTCTTCCTTAT | 22 ± 3 | <1 |
| PO2 | GAAGGGCTTCTTCCTTCT | 38 ± 4 | <1 |
| PO3 | GAAGGGCTTCTTCCTTGT | 19 ± 1 | |
| PO4 | GAAGGGCTTCTTCCTTTT | 17 ± 1 | |
| PO5 | GAAGGGCTTCTTCCTTAA | ||
| PO6 | GAAGGGCTTCTTCCTCAT | 24 ± 1 | |
| PO7 | GAAGGGCTTCTTCCTGAT | 29 ± 2 | |
| PO8 | GAAGGGCTTCTTCCTTAC | 32 ± 1 | |
| PO9 | GAAGGGCTTCTTCCTTAG | 47 ± 2 | |
| PO10 | AGTCTTGCATCGTCTTGA | ||
| PO11 | ATTCTGTGAAGTCCTTGC | ||
| PO12 | ATAAGGAAGAAGCCCTTC | 37 ± 4 | |
| PO13 | TTTTTTTTTTTTTTTTTT | 12 ± 1 | |
| PO14 | CCCCCCCCCCCCCCCCCC | >72 | |
| PO/PS1 | GAAGGGCTTCTTCCTTA(Ps)T | >100 | 15 ± 1 |
| PO/PS2 | GAAGGGCTTCTTCCTT(pS)AT | 23 ± 4 | <1 |
| PO/PS3 | GAAGGGCTTCTTCCT(pS)TAT | 25 ± 2 | <1 |
| PS1 | CGCTGAAGGGCTTCTTCCTTATTGAT | ||
| PS2 | ATCAATAAGGAAGAAGCCCTTCAGCG | ||
| PS3 | CGCTGAAGGGCTTTTGAACTCTGCTT | ||
| PS4 | AAGCAGAGTTCAAAAGCCCTTCAGCG | ||
| PS5 | CGCTGAAGGGCTTCTTCCTTATTGCG | ||
| PS6 | ATCAATAAGGAAGAAGCCCTTCAGAT | ||
Phosphodiester (PO1 TO PO14), chimeric phosphodiester/phosphorothioate (PO/PS1 to PO/PS3), and all-phosphorothioate (PS1 to PS6) ODNs used in this study. The position of the phosphorothioate linkage is indicated by “pS” in the chimeric ODN sequences. Half-lives of the full-length ODNs are given in hours and correspond to an incubation of the ODNs at a 25 μmol/L concentration and at 37°C in RPMI 1640 culture media containing 2.5% NHS or 10% FCS. Data are the mean of three experiments ± SD.
Effect of Phosphodiester and Phosphorothioate ODNs on BV173
| ODN . | 3′ End . | Cell Counts After 72 Hours . | ||
|---|---|---|---|---|
| . | . | (% of Control ± SD) . | ||
| . | . | 2.5% NHS . | 2.5% NHSΔ . | 10% FCS . |
| PO1 | TAT | 35 ± 8 | 105 ± 7 | 114 ± 12 |
| PO2 | TCT | 81 ± 5 | ||
| PO3 | TGT | 18 ± 5 | ||
| PO4 | TTT | 25 ± 2 | ||
| PO5 | TAA | 37 ± 3 | ||
| PO6 | CAT | 53 ± 5 | ||
| PO7 | GAT | 27 ± 10 | ||
| PO8 | TAC | 118 ± 13 | ||
| PO9 | TAG | 22 ± 8 | ||
| PO10 | TGA | 22 ± 6 | 97 ± 9 | |
| PO11 | TGC | 105 ± 7 | 92 ± 11 | |
| PO12 | TTC | 119 ± 13 | 101 ± 11 | 104 ± 4 |
| PO13 | TTT | 2 ± 2 | ||
| PO14 | CCC | 123 ± 1 | ||
| PO/PS1 | TTA(pS)T | 91 ± 14 | ||
| PO/PS2 | TT(pS)AT | 36 ± 6 | ||
| PO/PS3 | T(pS)TAT | 29 ± 4 | ||
| ODN . | 3′ End . | Cell Counts After 72 Hours . | ||
|---|---|---|---|---|
| . | . | (% of Control ± SD) . | ||
| . | . | 2.5% NHS . | 2.5% NHSΔ . | 10% FCS . |
| PO1 | TAT | 35 ± 8 | 105 ± 7 | 114 ± 12 |
| PO2 | TCT | 81 ± 5 | ||
| PO3 | TGT | 18 ± 5 | ||
| PO4 | TTT | 25 ± 2 | ||
| PO5 | TAA | 37 ± 3 | ||
| PO6 | CAT | 53 ± 5 | ||
| PO7 | GAT | 27 ± 10 | ||
| PO8 | TAC | 118 ± 13 | ||
| PO9 | TAG | 22 ± 8 | ||
| PO10 | TGA | 22 ± 6 | 97 ± 9 | |
| PO11 | TGC | 105 ± 7 | 92 ± 11 | |
| PO12 | TTC | 119 ± 13 | 101 ± 11 | 104 ± 4 |
| PO13 | TTT | 2 ± 2 | ||
| PO14 | CCC | 123 ± 1 | ||
| PO/PS1 | TTA(pS)T | 91 ± 14 | ||
| PO/PS2 | TT(pS)AT | 36 ± 6 | ||
| PO/PS3 | T(pS)TAT | 29 ± 4 | ||
BV173 cell viability expressed as a percentage of control cell counts 72 hours after a single addition of phosphodiester or chimeric ODN, at a concentration of 25 μmol/L, in RPMI 1640 containing 2.5% NHS, 2.5% NHS heat-treated at 60°C for 30 minutes (NHSΔ), or 10% FCS. Data are expressed as the mean of at least three experiments ± SD.
BV173 cell viability (expressed as a percentage of control cell counts) as a function of time (in hours) following a single addition of ODN at a concentration of 25 μmol/L, in RPMI 1640 containing 2.5% NHS. Data are expressed as the mean of at least three experiments ± SD.
BV173 cell viability (expressed as a percentage of control cell counts) as a function of time (in hours) following a single addition of ODN at a concentration of 25 μmol/L, in RPMI 1640 containing 2.5% NHS. Data are expressed as the mean of at least three experiments ± SD.
ODN chemistry and antiproliferative effect on BV173.To further investigate the relationship between the 3′ terminal sequences of the ODNs and their biological effect, we constructed the phosphodiester/phosphorothioate chimeric sequences PO/PS1 to PO/PS3 (Table 1). These sequences were identical to PO1, but contained a phosphorothioate linkage instead of a phosphodiester at one of the last three 3′ sugar linkages. At an ODN concentration of 25 μmol/L in culture medium containing 2.5% NHS, PO/PS2 and PO/PS3 were active (Table 2), as their corresponding sequence PO1. On the contrary, PO/PS1 was inactive: a single phosphorothioate chemical transformation of the last 3′ phosphodiester position of PO1 was sufficient to abolish its antiproliferative effect.
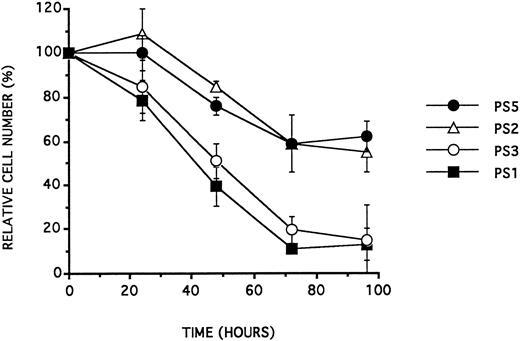
26-mer PS ODNs and antiproliferative effect on BV173.26-mer antisense all-phosphorothioate ODN analogues, which target the junctional regions of the BCR/ABL mRNA, were reported to inhibit the proliferation of BV17321 in RPMI 1640 containing 10% FCS. To analyze whether this effect could also be due to the 3′ base sequence of the ODNs only, we incubated BV173 cultured in the same conditions (10% FCS) with the antisense PS1 and PS3 (which target, respectively, the b2a2 or the b3a2 junctional region of BCR/ABL mRNA), with the corresponding sense PS2 and PS4 and with the control ODNs PS5 and PS6 presented in Table 1. PS5 and PS6 were synthesized with their sequence identical to PS1 or PS2, except for their last two 3′ bases: the last two bases of the antisense sequences PS1 and PS2 were transposed. Those experiments were performed at an ODN concentration of 10 μmol/L. As for phosphodiester ODNs, the antiproliferative effect of these ODNs depended on their 3′ sequence (Table 3 and see Fig 2).
Effect of Phosphorothioate ODNs on BV173
| ODN . | 3′ End . | Cell Counts After 72 Hours . | |
|---|---|---|---|
| . | . | (% of Control ± SD) . | |
| . | . | 2.5% NHS . | 10% FCS . |
| PS1 | GAT | 83 ± 10 | 11 ± 1 |
| PS2 | GCG | 84 ± 11 | 59 ± 13 |
| PS3 | CTT | 19 ± 6 | |
| PS4 | GCG | 59 ± 10 | |
| PS5 | GCG | 59 ± 1 | |
| PS6 | GAT | 18 ± 0 | |
| ODN . | 3′ End . | Cell Counts After 72 Hours . | |
|---|---|---|---|
| . | . | (% of Control ± SD) . | |
| . | . | 2.5% NHS . | 10% FCS . |
| PS1 | GAT | 83 ± 10 | 11 ± 1 |
| PS2 | GCG | 84 ± 11 | 59 ± 13 |
| PS3 | CTT | 19 ± 6 | |
| PS4 | GCG | 59 ± 10 | |
| PS5 | GCG | 59 ± 1 | |
| PS6 | GAT | 18 ± 0 | |
BV173 cell viability expressed as a percentage of control cell counts 72 hours after a single addition of phosphorothioate ODN at a concentration of 10 μmol/L, in RPMI 1640 containing 2.5% NHS or 10% FCS. Data are expressed as the mean of at least three experiments ± SD.
BV173 cell viability (expressed as a percentage of control cell counts) as a function of time (in hours) following a single addition of ODN at a concentration of 10 μmol/L, in RPMI 1640 containing 10.0% FCS. Data are expressed as the mean of at least three experiments ± SD.
BV173 cell viability (expressed as a percentage of control cell counts) as a function of time (in hours) following a single addition of ODN at a concentration of 10 μmol/L, in RPMI 1640 containing 10.0% FCS. Data are expressed as the mean of at least three experiments ± SD.
Culture medium and antiproliferative effect on BV173.To analyze the influence of the culture medium on the antiproliferative effect, we incubated BV173 with ODNs, in culture medium containing different sera.
With phosphodiester ODNs (Table 2), the cell growth inhibition induced by the active ODNs was only observed in culture media containing 2.5% NHS. In 10% FCS or 2.5% heat-treated NHS (60°C, 30 minutes), the effect was not observed. However, the homopolymer PO13 (but not PO14) was active in media containing 10% FCS.
With phosphorothioate ODNs (Table 3), the activity observed for PS1, PS3, and PS6 in media containing 10% FCS was abolished on replacement of 10% FCS with 2.5% NHS.
Degradation kinetics of ODNs in culture media.Phosphodiester and chimeric phosphodiester/phosphorothioate ODNs were incubated in culture media containing different sera. The stepwise hydrolysis of the ODN phosphate linkages by 3′ exonucleases was observed, as already reported.43-48 This degradation process was monitored and quantitated using ion exchange chromatography.
In comparison with medium containing 10% FCS, the 2.5% NHS containing medium was at least 10 times less active in terms of ODN 3′ hydrolysis (Table 1).
Heating of NHS before use (60°C, 30 minutes) drastically slowed down the degradation process: no degradation pattern of the ODNs was observed after 72 hours of incubation.
As already reported,47 the stability of the phosphodiester ODNs towards 3′ exonucleases present in NHS was dependent on their last 3′ phosphodiester linkage (Table 1).
The chimeric ODN, PO/PS1, was completely stable in 2.5% NHS containing media. In 10% FCS, an hydrolysis of the phosphorothioate linkage was observed. The half-life of PO/PS1 was about 15 hours. There was no difference between the two diastereoisomers of PO/PS1 (distinguishable using ion exchange chromatography) in terms of stability.
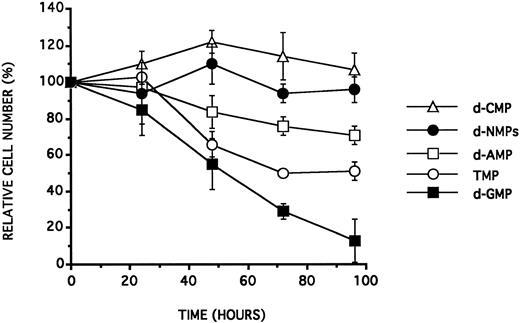
Toxicity of d-NMPs on BV173.The BV173 cell line, cultured in a 2.5% NHS containing medium, was incubated with increasing concentrations of d-NMPs. As illustrated in Table 4 and see Fig 3,10 at a concentration of at least 10 μmol/L, all the d-NMPs except d-CMP inhibited cell growth after 48 hours of incubation. The following decreasing order of effectiveness was found: d-GMP > TMP > d-AMP. The equimolar mixture of the four d-NMPs, at concentrations up to 100 μmol/L each, had no effect. Experiments with the simultaneous presence of two d-NMPs showed furthermore that a coincubation with an equimolar amount of d-CMP strongly diminished the antiproliferative effect of any other d-NMP.
Effect of d-NMPs on BV173
| d-NMP . | Concentration (μmol/L) . | Cell Counts After 72 Hours . | ||
|---|---|---|---|---|
| . | . | (% of Control ± SD) . | ||
| . | . | 2.5% NHS . | 2.5% NHSΔ . | 10% FCS . |
| d-AMP | 1 | 108 ± 18 | ||
| d-AMP | 10 | 80 ± 7 | ||
| d-AMP | 25 | 76 ± 5 | 90 ± 10 | 79 ± 3 |
| d-AMP | 100 | 54 ± 4 | ||
| d-CMP | 1 | 106 ± 6 | ||
| d-CMP | 10 | 99 ± 13 | ||
| d-CMP | 25 | 114 ± 13 | 102 ± 18 | 99 ± 8 |
| d-CMP | 100 | 119 ± 23 | ||
| d-GMP | 1 | 105 ± 7 | ||
| d-GMP | 10 | 37 ± 15 | ||
| d-GMP | 25 | 29 ± 4 | 95 ± 8 | 37 ± 5 |
| d-GMP | 100 | 14 ± 2 | ||
| TMP | 1 | 107 ± 10 | ||
| TMP | 10 | 71 ± 7 | ||
| TMP | 25 | 50 ± 1 | 97 ± 5 | 61 ± 13 |
| TMP | 100 | 41 ± 8 | ||
| d-NMPs | 100 (400) | 94 ± 5 | ||
| d-AMP+d-CMP | 25 (50) | 113 ± 15 | ||
| d-AMP+d-GMP | 25 (50) | 41 ± 11 | ||
| d-AMP+TMP | 25 (50) | 45 ± 7 | ||
| d-CMP+d-GMP | 25 (50) | 59 ± 9 | ||
| d-CMP+TMP | 25 (50) | 113 ± 21 | ||
| d-GMP+TMP | 25 (50) | 17 ± 5 | ||
| d-NMP . | Concentration (μmol/L) . | Cell Counts After 72 Hours . | ||
|---|---|---|---|---|
| . | . | (% of Control ± SD) . | ||
| . | . | 2.5% NHS . | 2.5% NHSΔ . | 10% FCS . |
| d-AMP | 1 | 108 ± 18 | ||
| d-AMP | 10 | 80 ± 7 | ||
| d-AMP | 25 | 76 ± 5 | 90 ± 10 | 79 ± 3 |
| d-AMP | 100 | 54 ± 4 | ||
| d-CMP | 1 | 106 ± 6 | ||
| d-CMP | 10 | 99 ± 13 | ||
| d-CMP | 25 | 114 ± 13 | 102 ± 18 | 99 ± 8 |
| d-CMP | 100 | 119 ± 23 | ||
| d-GMP | 1 | 105 ± 7 | ||
| d-GMP | 10 | 37 ± 15 | ||
| d-GMP | 25 | 29 ± 4 | 95 ± 8 | 37 ± 5 |
| d-GMP | 100 | 14 ± 2 | ||
| TMP | 1 | 107 ± 10 | ||
| TMP | 10 | 71 ± 7 | ||
| TMP | 25 | 50 ± 1 | 97 ± 5 | 61 ± 13 |
| TMP | 100 | 41 ± 8 | ||
| d-NMPs | 100 (400) | 94 ± 5 | ||
| d-AMP+d-CMP | 25 (50) | 113 ± 15 | ||
| d-AMP+d-GMP | 25 (50) | 41 ± 11 | ||
| d-AMP+TMP | 25 (50) | 45 ± 7 | ||
| d-CMP+d-GMP | 25 (50) | 59 ± 9 | ||
| d-CMP+TMP | 25 (50) | 113 ± 21 | ||
| d-GMP+TMP | 25 (50) | 17 ± 5 | ||
BV173 viable cell counts (expressed as a percentage of control cell counts) 72 hours after a single addition of d-NMP (or of d-NMP mixture), at concentrations from 1 to 400 μmol/L in RPMI 1640 containing 2.5% NHS, 2.5% NHS heat-treated at 60°C for 30 minutes (NHSΔ), or 10% FCS. Data are expressed as the mean of at least three experiments ± SD. The d-NMP mixtures are composed of equimolar amounts of each component. The total d-NMP concentration is indicated between brackets.
BV173 viable cell counts (expressed as a percentage of control cell counts) after a single addition of one of the four d-NMPs (d-AMP, d-CMP, d-GMP, and TMP) at a concentration of 25 μmol/L or of an equimolar (100 μmol/L of each) mixture of the four d-NMPs in RPMI 1640 supplemented with 2.5% NHS. Data are expressed as the mean of at least three experiments ± SD.
BV173 viable cell counts (expressed as a percentage of control cell counts) after a single addition of one of the four d-NMPs (d-AMP, d-CMP, d-GMP, and TMP) at a concentration of 25 μmol/L or of an equimolar (100 μmol/L of each) mixture of the four d-NMPs in RPMI 1640 supplemented with 2.5% NHS. Data are expressed as the mean of at least three experiments ± SD.
In FCS containing media, the results were identical to those obtained in NHS containing ones. On the contrary, in media containing 2.5% heat-treated NHS (60°C, 30 minutes), no antiproliferative effect was observed.
Antiproliferative effects on CD34 positive bone marrow cells.The ODN and the d-NMP induced antiproliferative effects were tested on fresh hematopoietic cells: we performed culture experiments (Table 5) on bone marrow cells, enriched in CD34+ cells, derived from healthy volunteers. Cell counts always displayed profiles similar to those observed, under the same experimental conditions, on BV173.
Effect of d-NMPs and ODNs on CD34+ Bone Marrow Cells
| d-NMP or ODN . | 3′ End . | Concentration (μmol/L) . | Cell Counts After 72 Hours . | |
|---|---|---|---|---|
| . | . | . | (% of Control ± SD) . | |
| . | . | . | 2.5% NHS . | 10% FCS . |
| d-AMP | 25 | 80 ± 2 | ||
| d-CMP | 25 | 106 ± 1 | ||
| d-GMP | 25 | 24 ± 6 | ||
| TMP | 25 | 53 ± 21 | ||
| d-NMP | 25 (100) | 92 ± 3 | ||
| PO1 | TAT | 25 | 42 ± 5 | |
| PO12 | TTC | 25 | 104 ± 3 | |
| PS1 | GAT | 10 | 38 ± 7 | |
| PS2 | GCG | 10 | 84 ± 5 | |
| PS3 | CTT | 10 | 30 ± 4 | |
| PS4 | GCG | 10 | 86 ± 11 | |
| PS5 | GCG | 10 | 83 ± 4 | |
| PS6 | GAT | 10 | 45 ± 1 | |
| d-NMP or ODN . | 3′ End . | Concentration (μmol/L) . | Cell Counts After 72 Hours . | |
|---|---|---|---|---|
| . | . | . | (% of Control ± SD) . | |
| . | . | . | 2.5% NHS . | 10% FCS . |
| d-AMP | 25 | 80 ± 2 | ||
| d-CMP | 25 | 106 ± 1 | ||
| d-GMP | 25 | 24 ± 6 | ||
| TMP | 25 | 53 ± 21 | ||
| d-NMP | 25 (100) | 92 ± 3 | ||
| PO1 | TAT | 25 | 42 ± 5 | |
| PO12 | TTC | 25 | 104 ± 3 | |
| PS1 | GAT | 10 | 38 ± 7 | |
| PS2 | GCG | 10 | 84 ± 5 | |
| PS3 | CTT | 10 | 30 ± 4 | |
| PS4 | GCG | 10 | 86 ± 11 | |
| PS5 | GCG | 10 | 83 ± 4 | |
| PS6 | GAT | 10 | 45 ± 1 | |
CD34+ bone marrow cell counts expressed as a percentage of control cells 72 hours after a single addition of d-NMP, d-NMP mixture or ODN, at concentrations from 25 to 100 μmol/L (for d-NMPs) or 10 to 25 μmol/L (for phosphorothioate or phosphodiester ODNs), in RPMI 1640 containing 2.5% NHS or 10% FCS. Data are expressed as the mean of three experiments (each on a different bone marrow) ± SD.
DISCUSSION
We have previously shown that phosphodiester ODNs with a “TAT” motif at their 3′ end inhibit the growth of normal and leukemic CD34+ hematopoietic cells as well as leukemic cell lines BV173 and KCL22. The results presented here further show that the “TAT” sequence is not the only one inducing an antiproliferative effect. All tested phosphodiester ODNs were active, except those containing a “C” at one of their two most terminal 3′ positions. These observations are also applicable to phosphorothioate ODNs. Considering that 50% of ODNs of random sequence are devoid of “C” at one of their two most terminal 3′ positions, 50% of random ODNs will therefore inhibit hematologic cell growth.
The absence of a “C” at one of the two most terminal 3′ positions of an ODN is necessary but not sufficient to induce the antiproliferative effect. The phosphodiester ODNs without “C” were only active in medium containing 2.5% NHS where their half-lives were of about 24 hours. When NHS was heat-treated, the antiproliferative effect and the ODN degradation process disappeared. The chimeric phosphodiester/phosphorothioate ODN, PO/PS1, containing only one phosphorothioate linkage (and no “C”) at its 3′ end, was inactive in 2.5% NHS, in which it was very stable (half-life >100 hours). The all-phosphorothioate ODNs were inactive and stable in 2.5% NHS. In 10% FCS, the phosphorothioate ODNs were partially hydrolyzed, with half-lives of about 24 hours,48 and, under such experimental conditions, all the ODNs without a “C” exerted an effect on the cells, as phosphodiester ODNs in 2.5% NHS. We conclude that the antiproliferative effect is only observed in culture media in which the ODNs are degraded by the 3′ exonucleases present in the sera. However, the absence of a “C” and the extracellular instability of ODNs do not sufficiently explain all our experimental data. In media containing 10% FCS, the phosphodiester ODNs, despite of half-lives shorter than 1 hour, were all inactive. The 18-mer poly-T homopolymer PO13 was an exception, with an half-life of a few minutes and an effect on cell growth. Thus, it appears (with the exception of the PO13 behavior) that the ODN degradation kinetics control the antiproliferative effect: to observe the antiproliferative effect, the ODNs have to be hydrolyzed but not too rapidly.
To explain this paradoxical effect, we analyzed the cytotoxicity of d-NMPs, the monomeric degradation products of the phosphodiester ODNs. At concentrations around 10 μmol/L, the cytotoxicity of the different d-NMPs was not identical. Not only d-CMP was inactive on the cells but it also suppressed (or diminished) the toxic effect of other d-NMPs. The mixture of the four d-NMPs, each at a concentration of 100 μmol/L (as a model of the d-NMP concentrations released by ODNs in 10% FCS), was not toxic. Similar observations were already published on hematologic cells or cell lines.50 51
We conclude that the antiproliferative activity of the phosphodiester (and most probably phosphorothioate) ODNs is due to the stepwise release of d-NMPs under the effect of serum exonucleases. The effect is restricted to experimental culture conditions in which the 3′ hydrolysis generates a significant amount of d-NMPs other than d-CMP.
All our experimental data regarding the phosphodiester or phosphodiester/phosphorothioate chimeric ODNs conforms with this explanation.
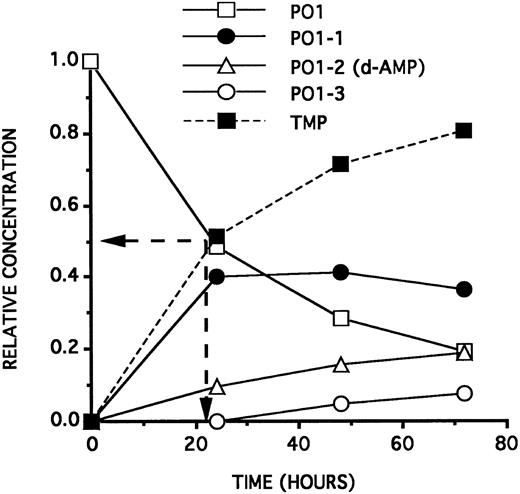
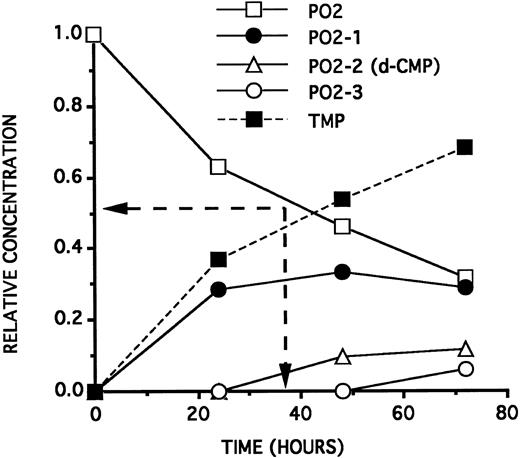
The toxic doses of d-NMPs on the cells are compatible with the d-NMP concentrations released by the ODNs as a function of time. As deduced from Fig 4,10 PO1, incubated at a concentration of 25 μmol/L in a culture medium containing 2.5% NHS, will generate by its 3′ hydrolysis, after 24 hours, a 12.5 μmol/L TMP concentration and a 1.0 μmol/L d-AMP concentration. Forty-eight hours later, an antiproliferative effect can be observed on the cells. On the contrary, PO2, incubated at a concentration of 25 μmol/L in a culture medium containing 2.5% NHS, will generate (see Fig 5) by its 3′ hydrolysis, after 24 hours, a 5.0 μmol/L TMP concentration and a d-CMP concentration lower than 1.0 μmol/L. Forty-eight hours later, no antiproliferative effect is observed on the cells. This low TMP concentration will not induce an inhibition of the cell growth 48 hours later.
Kinetics curves (relative ODN concentrations as a function of time) of the 3′ hydrolysis of a 25 μmol/L solution of PO1 on incubation at 37°C in RPMI 1640 containing 2.5% HS. PO1-1, PO1-2, and PO1-3 represent, respectively, the PO1 sequence minus 1, 2, and 3 deoxyribonucleotides at the 3′ end. PO1 half-life (22 hours) is indicated by the position of the two arrows. The dashed curve (symmetrical to the PO1 curve) indicates the theoretical TMP relative concentration generated by hydrolysis of the PO1 3′ last linkage. The PO1-2 curve represents also the maximal d-AMP relative concentration generated by hydrolysis of the PO1-1 3′ last position.
Kinetics curves (relative ODN concentrations as a function of time) of the 3′ hydrolysis of a 25 μmol/L solution of PO1 on incubation at 37°C in RPMI 1640 containing 2.5% HS. PO1-1, PO1-2, and PO1-3 represent, respectively, the PO1 sequence minus 1, 2, and 3 deoxyribonucleotides at the 3′ end. PO1 half-life (22 hours) is indicated by the position of the two arrows. The dashed curve (symmetrical to the PO1 curve) indicates the theoretical TMP relative concentration generated by hydrolysis of the PO1 3′ last linkage. The PO1-2 curve represents also the maximal d-AMP relative concentration generated by hydrolysis of the PO1-1 3′ last position.
Kinetics curves (relative ODN concentrations as a function of time) of the 3′ hydrolysis of a 25 μmol/L solution of PO2 on incubation at 37°C in RPMI 1640 containing 2.5% HS. PO2-1, PO2-2, and PO2-3 represent, respectively, the PO2 sequence minus 1, 2, and 3 deoxyribonucleotides at the 3′ end. PO2 half-life (38 hours) is indicated by the position of the two arrows. The dashed curve (symetrical to the PO2 curve) indicates the theoretical TMP relative concentration generated by hydrolysis of the PO2 3′ very last linkage. The PO2-2 curve represents also the maximal d-CMP relative concentration generated by hydrolysis of the PO2-1 3′ very last position.
Kinetics curves (relative ODN concentrations as a function of time) of the 3′ hydrolysis of a 25 μmol/L solution of PO2 on incubation at 37°C in RPMI 1640 containing 2.5% HS. PO2-1, PO2-2, and PO2-3 represent, respectively, the PO2 sequence minus 1, 2, and 3 deoxyribonucleotides at the 3′ end. PO2 half-life (38 hours) is indicated by the position of the two arrows. The dashed curve (symetrical to the PO2 curve) indicates the theoretical TMP relative concentration generated by hydrolysis of the PO2 3′ very last linkage. The PO2-2 curve represents also the maximal d-CMP relative concentration generated by hydrolysis of the PO2-1 3′ very last position.
In media containing 10% FCS, the unstable phosphodiester ODNs, with half-lives of about a few minutes, are inactive because of the presence of d-CMP in the d-NMP mixture resulting from their rapid 3′ hydrolysis. On the contrary, the homopolymer PO13 is still active in 10% FCS because of the toxicity of its monomer TMP and the absence of “C ” in its sequence.
In contrast to the behavior of PO/PS1, the chimeric ODNs PO/PS2 and PO/PS3 are active in 2.5% NHS because, unlike PO/PS1, they release TMP at a concentration of at least a 10 μmol/L after 24 hours of incubation.
It is noticeable that the type of the culture medium is also important for the activity of the d-NMPs: no effect is observed in heat-treated NHS containing media. We hypothesize that one or several metabolic products of the d-NMPs (such as deoxyribonucleosides), may be involved in the cytotoxicity and that the heat-treatment of NHS inactivates key enzymes of this metabolism. This point, under current investigation, is already strongly supported by several published data.52-59
The toxicity of 5′ phosphorothioate deoxyribonucleosides has still to be examined to fully show the extension of our conclusions to phosphorothioate ODNs.
The degradation kinetics of ODNs in various media has been and is still being studied by several groups.43-48 The putative cellular effects of the ODN degradation products have already been pointed out.49 The biological activity of d-NMPs on hematologic cells has been also reported in the literature.50-59 However, our work shows, for the first time, that the 3′ sequence of an ODN is responsible for its biological activity via its stepwise hydrolysis.
Different groups reported an antiproliferative effect of anti-BCR/ABL ODNs on leukemic cells and cell lines. This effect, sometimes reported as an antisense effect, was suspected by different groups to be not junction specific, not leukemia specific and even not ODN sequence specific. To show an antisense effect, the direct measurement of the target protein level, as compared with that of internal controls, is virtually mandatory. A selective downregulation of the BCR/ABL protein, in an experimental system consisting of plain incubation of cells with ODNs, would of course be an indication of a possible specific antisense effect. However, such a selective inhibition, in these experimental conditions, has not been shown and protein measurement are sometimes difficult to perform in cells dying because of the effect induced by ODNs. To tackle this confusing situation, we decided to use many control ODNs to investigate the antiproliferative effect. After analysis of the degradation kinetics of the ODNs under our cell culture conditions, we are now able to interpret this effect: this antiproliferative activity of anti-BCR/ABL ODNs on CML cells should be re-interpreted as an example of non-antisense, but sequence-specific, cellular toxicity. This antiproliferative effect of the ODNs is programmed in their 3′ sequences, but is only executed if their degradation generates an “active” d-NMP mixture in the culture medium. The sensitivity of normal CD34+ bone marrow cells shows that the mechanism of this effect is not leukemia-specific. Since our goal was to understand this mechanism, and see whether it applied also to normal cells, we have performed no dose-effect relationship on normal and leukemic CD34+ cells. Therefore, we may not exclude a difference of sensitivity to cytotoxic d-NMPs between normal and leukemic cells. As a result, we may not exclude that dNMPs could be used for bone marrow purging if there were a preferential toxic effect on malignant cells. Anyhow the use of phosphodiester or phosphorothioate ODNs for purging purposes is questionable, at least in experimental conditions under which d-NMPs are produced in the μmol/L concentration range by the extracellular degradation of the ODNs. Nevertheless, our results do not preclude the validity of other approaches using different methods to introduce ODNs inside the cells. The question of whether the BCR/ABL mRNA, could be a good target for an antisense approach in CML is still under study in several groups and deserves further investigations.60-63
Finally, our results also raise the question of whether other antisense experiments, using ODNs aimed at different targets, performed in conditions allowing extracellular production of d-NMPs, and leading to cell growth inhibition or, maybe, to other biological effects, should be reexamined using more stringent degradation controls. Our results clearly show that the commonly used control ODNs3 (sense, scrambled, random, mismatched) are not optimal controls for ODN degradation. When these control ODNs are hydrolyzed step by step during incubation time, they do not produce the same d-NMP mixture as the antisense ODN. This makes the use of at least another control ODN with the same 3′ sequence as the antisense mandatory in any experiment where a substantial extracellular 3′ degradation of the ODNs can be anticipated. Because the nuclease content of a particular serum is not constant but depends on the serum batch, on storage conditions and heat treatment,47 the ODN degradation processes should be examined, before performing any antisense experiment, for each particular incubation medium. The toxicity of the d-NMP mixtures produced at the concentrations predicted by the ODN degradation kinetics should then be assayed. The use of serum free or heat-treated serum media47 could minimize the degradation-induced toxicity.
ACKNOWLEDGMENT
We thank Drs Bernard Lebleu (Université de Montpellier I, CNRS, Montpellier, France), Nicole Straetmans (Université de Louvain, Hematologie, Brussels, Belgium), and Marc Lemaı̂tre (Eurogentec, Liège, Belgium) for critical reading of the manuscript.
Supported in part by the Fonds National de la Recherche Scientifique (FNRS, Brussels, Belgium) Grant No. 3.4576.94 and the EUROGENTEC Co. (Liège, Belgium). P.M. is a qualified investigator of the FNRS.
Address reprint requests to J.L. Vaerman, PhD, Laboratoire de Biologie Moléculaire Hématologique, Cliniques St Luc, Université Catholique de Louvain, Clos Chapelle-aux-champs 30/3052, 1200, Brussels, Belgium.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal