Abstract
Sequence variations in the Epstein-Barr virus (EBV) encoded latent membrane protein-1 (LMP-1) gene have been described in a Chinese nasopharyngeal carcinoma-derived isolate (CAO), and in viral isolates from various EBV-associated tumors. It has been suggested that these genetic changes, which include loss of a Xho I restriction site (position 169425) and a C-terminal 30-base pair (bp) deletion (position 168287-168256), define EBV genotypes associated with increased tumorigenicity or with disease among particular geographic populations. To determine the frequency of LMP-1 variations in European wild-type virus isolates, we sequenced the LMP1 promoter and gene in EBV from lymphoblastoid cell lines from healthy carriers and patients without EBV-associated disease. Sequence changes were often present, and defined at least four main groups of viral isolates, which we designate Groups A through D. The widespread prevalence of LMP-1 sequence variations, particularly the Xho I polymorphism and the 30-bp deletion, indicate that they cannot be used as simple markers for oncogenic viruses related to particular forms of EBV-associated tumor. Several of the structural changes detected occur, however, at sites where they may affect transcription, translation, or function of LMP-1. Future in vitro studies should aim to establish the functional importance of variations at these sites.
EPSTEIN-BARR VIRUS (EBV) encoded latent membrane protein 1 (LMP-1) is an integral membrane protein, which is a key molecule in EBV-induced cell transformation and shows in vitro oncogenic activity.1-8 LMP-1 may be important in the pathogenesis of EBV-associated tumors, particularly Hodgkin's disease (HD) and nasopharyngeal carcinoma NPC in which it is the only EBV protein with known oncogenic effect to be expressed.9,10 Several groups have suggested that sequence variations in LMP-1 define aggressive or geographically restricted oncogenic EBV genotypes.11-14 Hu et al11 described a nude mouse-passaged Chinese NPC tumor designated CAO that showed structural variations in the LMP-1 gene compared to the EBV laboratory prototype B95.8, consisting of 46 and 52 single base mutations in the LMP-1 promoter region and gene, respectively, a C-terminal 30-base pair (bp) deletion, and deletion of a 15-bp unique sequence with insertion of three 33-bp repeats in the repeat region.11 Transfection studies suggested that the CAO-LMP-1 gene and C1510, a similar variant reported by Chen et al,13 were more tumorigenic than the B95.8-LMP-1 gene.12,13 Both these LMP-1 variants have a G → T mutation (169425) resulting in loss of an Xho I restriction site. This mutation is significantly more frequent in EBV isolates from cases of Chinese compared with African NPC,11 and in isolates from Taiwanese NPC compared with samples from Taiwanese healthy controls.14
Recently, LMP-1 variants have been described in isolates from various EBV associated diseases from different geographical areas.15-20 In particular, the CAO-LMP-1–like C-terminal 30-bp deletion has been detected together with flanking mutations in about 30% of European HD17,20 (possibly associated with more aggressive disease15 ), 65% of European peripheral T-cell lymphomas (PTL), 100% of Malaysian PTLs, 76% of acquired immune deficiency (AIDS)-related lymphomas, and 100% of AIDS-related HD.15,17-20 However, the significance of these findings is hard to assess, as the frequency of the mutations in wild-type EBV isolates from asymptomatic EBV seropositive individuals has not been investigated. Furthermore, it is unclear which of the mutations found in the CAO-LMP-1 gene and promoter are associated with the Xho I restriction site loss and which with the 30-bp deletion, since these changes appear to occur independently.16 This makes it difficult to compare the results reported from Asian and Western countries.11 13-20 In this study we have investigated the frequency of sequence variations in LMP-1 in EBV isolates from healthy individuals and asymptomatic (pre and post) bone marrow transplant patients.
MATERIALS AND METHODS
Lymphoblastoid cell lines (LCLs) were established at the Department of Clinical and Tumor Immunology, Dr Daniel den Hoed Kliniek, Rotterdam, The Netherlands, and have previously been used for the investigation of EBNO types.21 EBV isolates (n = 62) were derived from LCLs from 34 European individuals (23 Dutch, 10 Swedish, 1 German), comprising 11 healthy EBV carriers and 23 allogenic bone marrow transplant recipients (6 pretransplantation and 17 posttransplantation) without overt EBV-related disease (see Table 1). In two patients, LCLs were established both before and after transplantation. In eight cases, two or more LCLs were established from the same, or serial blood samples, each individual showing consistent carriage of the same EBNO type.21 For polymerase chain reaction (PCR) analysis, cells were digested with proteinase-K (200 μg/mL) overnight at 55°C.
Overview of the 62 EBV Isolates in LCLs From 34 Individuals
| Group . | Donor No. . | LCL No. . | Xho I Site Present* . | Xho I Sequence† . | LMP-1 30-bp Deletion‡ . | LMP-1 Repeatsρ . | Additional Single Base Mutations†† . |
|---|---|---|---|---|---|---|---|
| A | 6 | 6 | yes | CTCGAG | no | 3+ | A 168839 G: no |
| TG 168216-15 GA: S369D | |||||||
| A | 10 | 10 | yes | CTCGAG | no | 3+ | G169334 C: W48C |
| C 168661 A: N220K | |||||||
| A | 11 | 11 | yes | CTCGAG | no | 3+ | T 169051 A: C116S |
| A | 12 | 12-13 | yes | CTCGAG | no | 3+ | A 169015 C: no |
| A | 33 | 61 | yes | CTCGAG | no | 3+ | G 169358 C: W39C |
| A 169207 C: I90L | |||||||
| A | 32 | 60 | yes | CTCGAG | no | 3+ | C 168964 T: no |
| A | 17 | 28-37 | yes | CTCGAG | no | 3+ | |
| A | 1 | 1 | yes | CTCGAG | no | 3+ | |
| A | 5 | 5 | yes | CTCGAG | no | 3+ | |
| A | 19 | 40-44∥ | yes | CTCGAG | no | 3+ | |
| A | 23 | 52 | yes | CTCGAG | no | 3+ | |
| A | 24 | 53 | yes | CTCGAG | no | 3+ | |
| A | 25 | 54 | yes | ND | no | 3+ | |
| A | 15 | 17 | yes | CTCGAG | ND | 3+ | |
| B | 8 | 8 | yes | CTCGAG | no | 6 | G 168653 T: R223I |
| A 169220 C: no | |||||||
| B | 9 | 9 | yes | CTCGAG | no | 6 | None |
| B | 27 | 56 | yes | ND | no | 3 | |
| B | 31 | 59 | yes | CTCGAG | no | 4 | |
| C | 2 | 2 | yes | CTCGAG | yes | 4 | C 169096 A: H101N |
| G 168658 C: E 221D | |||||||
| C | 3 | 3** | yes | CTCGAG | yes | 5 | C 168964 T: no |
| G 168658 C: E 221D | |||||||
| C | 4 | 4 | yes | CTCGAG | yes | 6 | G 169398 T: G 26V |
| G 168658 C: E 221D | |||||||
| C | 7 | 7 | yes | CTCGAG | yes | 7 | None |
| C | 13 | 14-15 | yes | CTCGAG | yes | 4 | See results |
| C | 14 | 16 | yes | CTCGAG | yes | 5 | C 168964 T: no |
| 21-27¶ | G 168658 C: E 221D | ||||||
| C | 30 | 58 | yes | CTCGAG | yes | 4 | C 168192 A: H377N |
| C | 20 | 45-47 | yes | CTCGAG | yes | 5 | |
| C | 21 | 48-49 | yes | CTCGAG | yes | 5 | |
| D | 26 | 55 | no | CTCTAC | no | 3 | G 168658 C: E221D |
| T 169015 G: no | |||||||
| C 168192 A: H377N | |||||||
| D | 29 | 57 | no | CTCTAC | no | 4 | G 168693 C: D214H |
| A 168191 G: H377R | |||||||
| D | 28 | 19,20 | no | CTCTAC | no | 3 | T 168211 C: no |
| C 168640 G: no | |||||||
| A 168191 G: H377R | |||||||
| D | 34 | 62 | no | CTCTAC | no | 3 | G 168658 C: E221D |
| C 168192 A: H377N | |||||||
| D | 18 | 38-39 | no | CTCTAC | no | 4 | |
| D | 22 | 51 | no | CTCTAC | no | 4 | |
| Not grouped | 16 | 18,50# | no | CTGGAC | no | 3+ |
| Group . | Donor No. . | LCL No. . | Xho I Site Present* . | Xho I Sequence† . | LMP-1 30-bp Deletion‡ . | LMP-1 Repeatsρ . | Additional Single Base Mutations†† . |
|---|---|---|---|---|---|---|---|
| A | 6 | 6 | yes | CTCGAG | no | 3+ | A 168839 G: no |
| TG 168216-15 GA: S369D | |||||||
| A | 10 | 10 | yes | CTCGAG | no | 3+ | G169334 C: W48C |
| C 168661 A: N220K | |||||||
| A | 11 | 11 | yes | CTCGAG | no | 3+ | T 169051 A: C116S |
| A | 12 | 12-13 | yes | CTCGAG | no | 3+ | A 169015 C: no |
| A | 33 | 61 | yes | CTCGAG | no | 3+ | G 169358 C: W39C |
| A 169207 C: I90L | |||||||
| A | 32 | 60 | yes | CTCGAG | no | 3+ | C 168964 T: no |
| A | 17 | 28-37 | yes | CTCGAG | no | 3+ | |
| A | 1 | 1 | yes | CTCGAG | no | 3+ | |
| A | 5 | 5 | yes | CTCGAG | no | 3+ | |
| A | 19 | 40-44∥ | yes | CTCGAG | no | 3+ | |
| A | 23 | 52 | yes | CTCGAG | no | 3+ | |
| A | 24 | 53 | yes | CTCGAG | no | 3+ | |
| A | 25 | 54 | yes | ND | no | 3+ | |
| A | 15 | 17 | yes | CTCGAG | ND | 3+ | |
| B | 8 | 8 | yes | CTCGAG | no | 6 | G 168653 T: R223I |
| A 169220 C: no | |||||||
| B | 9 | 9 | yes | CTCGAG | no | 6 | None |
| B | 27 | 56 | yes | ND | no | 3 | |
| B | 31 | 59 | yes | CTCGAG | no | 4 | |
| C | 2 | 2 | yes | CTCGAG | yes | 4 | C 169096 A: H101N |
| G 168658 C: E 221D | |||||||
| C | 3 | 3** | yes | CTCGAG | yes | 5 | C 168964 T: no |
| G 168658 C: E 221D | |||||||
| C | 4 | 4 | yes | CTCGAG | yes | 6 | G 169398 T: G 26V |
| G 168658 C: E 221D | |||||||
| C | 7 | 7 | yes | CTCGAG | yes | 7 | None |
| C | 13 | 14-15 | yes | CTCGAG | yes | 4 | See results |
| C | 14 | 16 | yes | CTCGAG | yes | 5 | C 168964 T: no |
| 21-27¶ | G 168658 C: E 221D | ||||||
| C | 30 | 58 | yes | CTCGAG | yes | 4 | C 168192 A: H377N |
| C | 20 | 45-47 | yes | CTCGAG | yes | 5 | |
| C | 21 | 48-49 | yes | CTCGAG | yes | 5 | |
| D | 26 | 55 | no | CTCTAC | no | 3 | G 168658 C: E221D |
| T 169015 G: no | |||||||
| C 168192 A: H377N | |||||||
| D | 29 | 57 | no | CTCTAC | no | 4 | G 168693 C: D214H |
| A 168191 G: H377R | |||||||
| D | 28 | 19,20 | no | CTCTAC | no | 3 | T 168211 C: no |
| C 168640 G: no | |||||||
| A 168191 G: H377R | |||||||
| D | 34 | 62 | no | CTCTAC | no | 3 | G 168658 C: E221D |
| C 168192 A: H377N | |||||||
| D | 18 | 38-39 | no | CTCTAC | no | 4 | |
| D | 22 | 51 | no | CTCTAC | no | 4 | |
| Not grouped | 16 | 18,50# | no | CTGGAC | no | 3+ |
Shading identifies donors from whom isolates have been sequenced from position 169835 (−320 compared to transcription start of LMP-1) to position 168125 (downstream to the LMP-1 stop codon). In the rest of the cases a smaller part of the promoter and only selected parts of the gene were sequenced (see Materials and Methods). LCL no. 1 to 11: LCLs from normal donors; 12 to 20: LCLs from pretransplant patients; 21 to 62: LCLs from posttransplant patients; *: Detection of the Xho I restriction site (C↓TCGAG: 169428-169423) on the EBV genome (according to Baer et al33 ); †: The sequencing result at the Xho I restriction site (CTCGAG: 169428-169423); underlining indicates mutations compared to B95.8; ‡: Presence or absence of the 30-bp deletion (position: 168287-168256); ρ: Number of 33-bp repeats in the repeat region of the LMP-1 gene (position: 168555-168400), 3+ indicates a repeat region identical to B95.8; ∥, ¶, #: indicate LCLs established from the same individual from serial blood samples; **: This EBV isolate was EBV subtype B, the rest were subtype A. ††Additional single base mutations detected and eventual amino acid changes.
Abbreviation: ND, not done.
EBV subtyping was done at the EBNA-2 locus in all 62 LCLs, using primers and PCR conditions described previously.22 The LMP-1 gene was analyzed in all 62 LCLs as follows: A 286-bp or 316-bp product (depending on presence or absence of the 30-bp deletion) was amplified and sequenced using primer pair LMP9/LMP11.23 Presence of the Xho I restriction site (position 169428-23) was analyzed using primer pair LMP123/LMP-PRO2 (AACAGTAGCGCCAAGAGCAG:169362-81). Following digestion with Xho I (Pharmacia Biotech, Weiterstadt, Germany) gel-electrophoresis showed two bands (46 bp and 67 bp) or one band (113 bp) depending on the presence or absence of the restriction site, respectively. The LMP-1 gene repeat region (position: 168555-168400) was analyzed using primer pair LMP-repeat5 (TCCCTCCCGCACCCTCAACAACAAGC:168723-01)/LMP-repeat3 (ACCGCCGCCACCGTCTGTCATC:168286-307). PCR product from the repeat region of isolates in which the whole LMP-1 gene was sequenced (see below), was used as control. Part of the LMP-1 promoter (−237 − +41, relative to transcription start) was amplified and sequenced using primers LMP-PRO1 (CTTTACCACCGCATTCCCAC:169752-33)/PRO2 in 45 isolates. At least one isolate was analyzed from each individual. Twenty of the 45 isolates were derived from the seven different individuals from whom two or more isolates had been established. This was done to confirm the results of the Xho I restriction assay and to further compare the LMP-1 gene in LCLs established from different individuals and those established from the same person. Based on the results of these analyses, 20 isolates from 20 individuals were selected for sequence analysis of a larger part of the promoter (−320 − +41) and the entire LMP-1 gene. These comprised five cases that had lost the Xho I restriction site (LCL no.: 18, 53, 55, 57, 62), seven cases with the 30-bp deletion (LCL no.: 2, 3, 7, 14, 16, 49, 58) and eight cases that had retained the Xho I restriction site and did not contain the 30-bp deletion (LCL no.: 5, 6, 8, 9, 10, 11, 60, 61). In these isolates, sequence analysis was performed from position 169835 (−320 compared to transcription start) to position 168125, downstream to the stop-codon of the LMP-1 gene. PCR product for sequence analysis of the promoter region and the entire LMP-1 gene was amplified using three sets of primers: Pro-0 (GCCGCCAACGACCTCCCAA:169875-57)/Pro-2, primers LMP-1/LMP-723 and primers LMP-5/LMP-11.23 The primers were selected to ensure overlapping PCR products for sequence analysis. In cases where PCR product was to be used for sequence analysis, two PCR reactions were performed for each primer set, one with a biotinylated upstream primer and one with a biotinylated downstream primer. When using the primer pairs Pro-0/Pro-2, LMPrepeat5/repeat3, and LMP-5/LMP-11 a standard PCR mix was used (Perkin Elmer-Cetus, Norwalk, CA), except for 0.2 mmol/L of deaza-GTP (Pharmacia Biotech) or 5 μL of dimethyl sulfoxide (DMSO). The template was denaturated at 98°C for 5 minutes. Subsequently, five cycles with an annealing temperature of 65°C, five cycles at 63°C, 25 cycles at 60°C, and five cycles at 57°C were performed. In all cycles the denaturation temperature was 94°C and the extension temperature was 74°C. The PCR conditions when using the other primer pairs were similar to those recently described.17 Bidirectional solid-phase dideoxy-sequencing was performed as previously described17 using the PRISM Sequenase Terminator Single-Stranded DNA Sequencing Kit (Applied Biosystems, Foster City, CA). Sequence analysis was performed on the semiautomated ABI 373A sequencer (Applied Biosystems). The PCR product generated by the primer pairs LMP-1/LMP-7 and LMP-5/LMP-11 necessitated use of additional sequencing primers LMP3,23LMP4A (AGATCTAACATTCCCTAGGA: 168938-19),LMP-8A (TAGGCGCACCTGGAGGTGGT: 168596-77), and LMP-9.23 These were placed 200 to 350 bp apart on both the sense and the antisense strand to ensure at least one readable sequence in both directions at any position in the analyzed DNA strands.
RESULTS
The 62 LCLs are listed according to the 34 individuals from whom they were established (Table 1). Comparison of the sequence results and the size of the repeat region showed that all LCLs established from the same individual contained EBV strains with identical sequences in the studied segments of the LMP-1 gene (data not shown) and with identical repeat regions (Table 1). The Xho I restriction site C ↓ TCGAG (position: 169428-169423) was retained in isolates from 27 of the 34 individuals (79%). Sequence analysis confirmed that the Xho I restriction site was only lost in cases in which mutations in the recognition sequence were found (Table 1). In six of the LCLs the Xho I polymorphism was due to a G → T mutation (169425). Two LCLs (18 and 48 from donor 16) had lost the restriction site because of a C → G mutation (169426) (Table 1). The 30-bp deletion (position: 168287-168256) was found in isolates from 9 of the 34 donors (26%) (Table 1). No deletions of any other size were detected in any of the cases. The size of the repeat region varied from three 33-bp repeats to seven 33-bp repeats. The isolates that had lost the Xho I restriction site due to the mutation G → T (169425) and the isolates with the 30-bp deletion all contained a repeat region different from B95.8. The two isolates (18 and 48) with the Xho I restriction site loss due to the mutation C → G (169426) and 14 of the 18 isolates which had retained the Xho I restriction site and did not contain the 30-bp deletion, had a repeat region similar to B95.8. The remaining four of these 18 isolates had a repeat region different from B95.8.
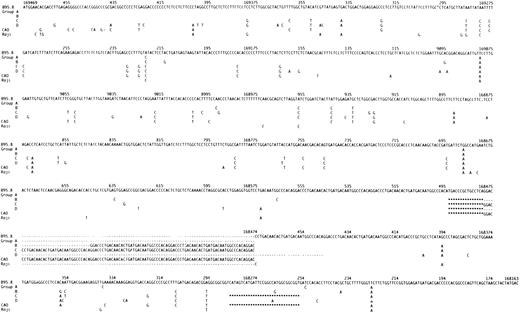
The sequence variation in the LMP-1 gene and promoter in the 20 fully sequenced isolates (with the exception of the isolate from LCL no. 18) consistently defined four groups (designated Groups A, B, C and D) (Figs 1, 2, and 3; Tables 1 and 2). Group A consisted of six isolates, with six shared single base mutations in the gene (Fig 1) and a single mutation in the promoter (C → T :169673) (data not shown), but which were otherwise identical to B95.8. Group B included two isolates with four mutations in the promoter (C → T: 169673; A → G: 169628; G → T: 169590; C → T: 169574) (data not shown), 19 identical single base mutations in the gene (Fig 1), the 15-bp deletion, and six 33-bp repeats, but without the 30-bp deletion (Fig 1 and Table 1). Group C consisted of 7 isolates. These were characterized by few mutations in the promoter (C → T: 169673 (all cases); 169686: deletion of a C (three cases); G → C: 169748 (one case); G → A: 169748 (two cases), data not shown), and 44 identical single base mutations in the gene (Fig 1). All contained the 15-bp deletion and the 30-bp deletion, had an insertion of one nucleotide (T) in intron two at position 168973 (data not shown) and had from four to seven 33-bp repeats in the C-terminus (Table 1). In addition, four isolates had an insertion of one nucleotide (C) and two isolates an insertion of two nucleotides (CC) in the first intron at position 169181 (data not shown). One of the LCLs (no. 14) contained an isolate with an additional 7-bp deletion and a single bp insertion from position 169469 to 169463 resulting in a change of the first amino acid after M from E to D (both negatively charged) and a deletion of the two following amino acids H and D, overall resulting in no change in charge (data not shown). Group D included 4 of the 5 isolates, which had lost the Xho I restriction site. This loss resulted from the single base mutation G → T (168925) (LCLs no.: 53, 55, 57, 62). The LMP-1 gene in these viruses was characterized by 35 identical mutations in the promoter region (Fig 2), 66 identical single base mutations in the gene (Fig 1) and an insertion of one nucleotide (T) in the second intron at position 168973 (data not shown). All isolates differed from B95.8 in having either 3 or 4 perfect 33-bp repeats. None of the Group D isolates contained the 30-bp deletion.
The nucleotide sequence of LMP-1 in the EBV strain B95.8 compared with our sequencing data for wild-type isolates, designated Groups A through D according to the pattern of mutations, and with CAO and Raji11 for comparison. Most of the isolates contained occasional additional single base mutations (see Table 1). For Groups A through D, CAO and Raji only differences compared to B95.8 are shown. Nucleotide numbering is according to Baer et al.34 Deletions are indicated with (***); the repeat region is indicated with (- - -).The number of 33-bp repeats varied within Groups B through D. A representative repeat region is shown for each group.
The nucleotide sequence of LMP-1 in the EBV strain B95.8 compared with our sequencing data for wild-type isolates, designated Groups A through D according to the pattern of mutations, and with CAO and Raji11 for comparison. Most of the isolates contained occasional additional single base mutations (see Table 1). For Groups A through D, CAO and Raji only differences compared to B95.8 are shown. Nucleotide numbering is according to Baer et al.34 Deletions are indicated with (***); the repeat region is indicated with (- - -).The number of 33-bp repeats varied within Groups B through D. A representative repeat region is shown for each group.
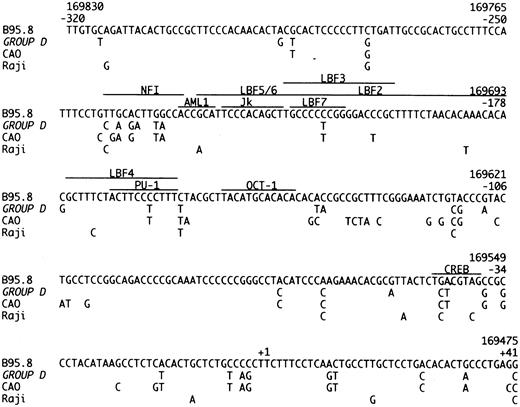
The promoter region of the LMP-1 gene, from position −320 to +41 relative to transcription start. The sequence of B95.8 is compared to our Group D cases, CAO and Raji.11 The recognition sequences of the shown transcription factors are marked with a line.35-37
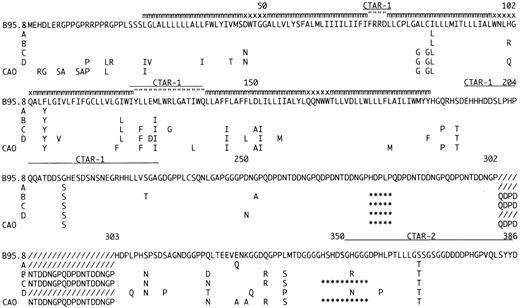
Amino acid sequence of LMP-1 in the sequenced EBV strains compared to B95.834 and CAO.11 The EBV strains are designated Groups A through D according to the pattern of mutations. Many of the isolates contained occasional additional amino acid mutations (data not shown). For Groups A through D and CAO only differences compared to B95.8 are shown. The positions of the carboxy-terminal activation region-1 (CTAR-1) and CTAR-2 are indicated by (__); deletions are indicated by (***); additional 33-bp repeats compared to B95.8 by (///); amino acids located within the plasma-membrane by (mmm); in the intra-cytoplasmatic part of the loops by (” ” ”); and on the outer surface by (xxx). The number of 33-bp repeats varied within Groups B through D. A representative repeat region is shown for each group.
Amino acid sequence of LMP-1 in the sequenced EBV strains compared to B95.834 and CAO.11 The EBV strains are designated Groups A through D according to the pattern of mutations. Many of the isolates contained occasional additional amino acid mutations (data not shown). For Groups A through D and CAO only differences compared to B95.8 are shown. The positions of the carboxy-terminal activation region-1 (CTAR-1) and CTAR-2 are indicated by (__); deletions are indicated by (***); additional 33-bp repeats compared to B95.8 by (///); amino acids located within the plasma-membrane by (mmm); in the intra-cytoplasmatic part of the loops by (” ” ”); and on the outer surface by (xxx). The number of 33-bp repeats varied within Groups B through D. A representative repeat region is shown for each group.
Comparison of the Number of Single Base Mutations, Amino Acid Changes, and Amino Acid Substitutions Resulting in a Change in Charge in the LMP-1 Gene in Virus Isolates From Groups A Through D
| Group . | A . | B . | C . | D . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | SBM . | AAC . | CIC . | SBM . | AAC . | CIC . | SBM . | AAC . | CIC . | SBM . | AAC . | CIC . |
| A | 6 | 5 | 1 | 4 | 4 | 0 | 4 | 4 | 0 | 4 | 4 | 0 |
| B | 4 | 4 | 0 | 19 | 14 | 2 | 10 | 7 | 1 | 10 | 7 | 1 |
| C | 4 | 4 | 0 | 10 | 7 | 1 | 44 | 20 | 4 | 33 | 14 | 1 |
| D | 4 | 4 | 0 | 10 | 7 | 1 | 33 | 14 | 1 | 66 | 35 | 8 |
| Group . | A . | B . | C . | D . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | SBM . | AAC . | CIC . | SBM . | AAC . | CIC . | SBM . | AAC . | CIC . | SBM . | AAC . | CIC . |
| A | 6 | 5 | 1 | 4 | 4 | 0 | 4 | 4 | 0 | 4 | 4 | 0 |
| B | 4 | 4 | 0 | 19 | 14 | 2 | 10 | 7 | 1 | 10 | 7 | 1 |
| C | 4 | 4 | 0 | 10 | 7 | 1 | 44 | 20 | 4 | 33 | 14 | 1 |
| D | 4 | 4 | 0 | 10 | 7 | 1 | 33 | 14 | 1 | 66 | 35 | 8 |
Groups A, B, C, and D refer to groups defined in Results. To use the table: Reading vertically and horizontally for the same group, it is possible to see the total number of single base mutations (SBM), amino acid changes (AAC), and amino acid changes which result in change of charge (CIC) in that group (eg, Group C isolates have 44 SBM). Comparing two separate groups shows the number of changes common for the two groups (eg, Groups C and D isolates have 33 SBM, 14 AAC, and 1 CIC in common).
Isolates could be allocated to Groups A through D on the basis of loss of the Xho I restriction site due to the mutation G → T (169425), changes in the repeat region and detection of the 30-bp deletion (Table 1). Isolate 18 was, apart from few single base mutations, identical to B95.8. However, it had lost the Xho I restriction site due to the C → G mutation (168926) and would have been falsely allocated to Group D if the restriction site had not been sequenced. We chose therefore not to group this isolate. The fully sequenced EBV isolates contained occasional single base mutations in addition to those shared in Groups A through D (Table 1). The degree of identity of the single base mutations between the groups and their effect on the amino acid sequence are shown in Figs 1 and 3, and in Table 2.
Using loss of the Xho I restriction site, detection of the 30-bp deletion and configuration of the repeat region the partly sequenced cases could each be allocated to the different groups. The total number of isolates in each group was then: Group A: 14/34 (41%); Group B: 4/34 (12%); Group C: 9/34 (26%); Group D: 6/34 (18%). One case (donor 16) was not grouped (3%) (Table 1).
DISCUSSION
In vitro studies have suggested that the NPC-derived CAO LMP-1 variant is more tumorigenic and less immunogenic than the laboratory prototype B95.8 LMP-1.12,13,24 These findings have prompted a search for CAO-associated genetic changes in EBV isolates from a variety of EBV-associated tumors15-20 and led to claims that these changes are associated with aggressive forms of HD and non-Hodgkin's lymphoma in both HIV-positive and -negative patients.17-20 Furthermore, Li et al25 have recently shown that introduction of the CAO-associated 30-bp deletion into B95.8-LMP-1 increases the transforming capacity and toxicity of the protein in Balb/3T3 cells, and renders these cells tumorigenic in nude mice.
The possible identification of EBV variants with increased oncogenicity has excited great interest, particularly as it may explain some of the striking epidemiological features of EBV infection and oncogenesis in different geographic populations. However, little is known about the normal distribution of variant LMP-1 in healthy individuals and our study was designed to provide this information.
Overall, our European wild-type isolates showed a high degree of conservation of the LMP-1 gene compared with the B95.8. However, we frequently identified a range of sequence variations (including the majority of those described in CAO-LMP-1) indicating that there is no simple association of these changes with EBV-associated tumors. LMP-1 sequence variants defined four main groups of wild-type isolates, which we have designated Groups A through D. The mutations defining each group are shown in Fig 1. Most isolates contained occasional additional mutations (Table 1). Full-length sequence analyses of wild-type viruses from other geographic regions may add to the number of groups.
The Group A viruses appear to be a substrain of B 95.8 with six single base mutations in the gene and a single mutation in the promoter. Group B isolates show an increased number of mutations (four in the promoter and 19 in the gene) together with the 15-bp deletion and a varying number of 33-bp repeats. Group C viruses are defined by the 30-bp deletion (Table 1, Fig 1). Of the 44 single base mutations found in the group, 35 (79%) were shared with CAO-LMP-111 (Fig 1), 36 (81%) shared with Clone151013 and 11 (25%) shared with Raji.11 We previously identified the 30-bp deletion in EBV isolates from 3/9 cases of infectious mononucleosis (IM),17 and the present study confirms that this change can be detected in the absence of overt EBV-associated disease. The frequency of the deletion in our material is similar to that found by us in European HD (approximately 30%).17 The 30-bp deletion has previously been associated with a variety of EBV-related tumors15-20 and our preliminary data also show an increased frequency of the deletion in isolates from Western NPC cases (76% of cases; Sandvej, Zhou, Hamilton-Dutoit, unpublished data, May 1995). We have previously found that EBV-subtype B isolates are often associated with the 30-bp deletion17 and the EBV-subtype B isolate included in this study also contained this deletion (Table 1). The increased frequency of the 30-bp deletion in AIDS-related lymphomas may partly reflect the presence of subtype B virus which is relatively more common in these tumors.26 However, this does not explain our findings in PTL and NPC where the increased frequency of the 30-bp deletion persists when EBV-subtype B cases are excluded (unpublished observations). Two of the amino acid changes consistently found together in Group C, but not detected in the CAO-LMP-1 may be important in explaining the apparent discordant data concerning tumorigenicity of the LMP-1 30-bp deletion variants. Firstly, there is substitution of the positively charged arginine132 with glycine (Fig 3). Huen et al27 have shown that this arginine is located in an important effector region of the LMP-1 protein (designated CTAR-1) and that substituting it with glutamic acid reduces the relative induction of surface markers CD54 and CD40 by 30% to 40% as compared with the B.95.8 LMP-1.27 Secondly, substitution of serine309 with asparagine is found in all isolates, with the exception of Group A variants (Fig 3). Moorthy and Thorley-Lawson28 have reported that substitution of serine309 creates an apparently less stable protein prone to degradation. The mutation found in our isolates may have a similar effect on LMP-1, thereby decreasing the half-life of the protein. The combined effect of these two amino acid changes could counterbalance any eventual increased tumorigenicity associated with introducing the 30-bp deletion.
Group D isolates are defined by loss of the Xho I restriction site due to the G → T (169425) mutation (Table 1). This restriction site polymorphism has been reported in NPC-derived isolates from Asia and Alaska.11,13,14,17 Group D isolates were further characterized by 35 identical single base mutations in the promoter region (Fig 2) and 66 identical single base mutations in the gene (Fig 1). CAO-LMP-111 show 74% homology (Fig 2), clone 151013 show 70% homology, and Raji11 show 20% homology (Fig 2) in the promoter mutations. Interestingly, most of the specific promoter mutations we found occurred at the position of, or adjacent to, specific promoter mutations in the CAO-LMP-1 gene suggesting these may be hotspots (Fig 2). The functional relevance of the promoter mutations is unknown. However, some of the mutational sites are of interest. The C → T (169722) mutation (−207) (Fig 2) removes a methylation site [CCGG at position 169722-169719 (−207 to −204)] thought to play a part in LMP-1 transcription control.29 Furthermore, mutations identical to those found in our Group D isolates in the recognition sequence of the CREB transcription factor (−45 to −39) (Fig 2)30 have been shown to decrease the LMP-1 promoter activity threefold to ninefold.25 30 This suggests that about 20% of wild-type EBV isolates (Group D) in this study contain a possibly weaker LMP-1 promoter. Interestingly, our preliminary studies suggest that Group D is underrepresented in EBV-associated HD (unpublished data). Isolate 18 had lost the Xho I restriction site but due to the C → G mutation (169426) and did not contain the Group D promoter or gene mutations. Thus, isolates showing Xho I restriction polymorphism should be sequenced at the restriction site before being allocated to Group D. Mutations were also found in the NFI transcription factor recognition sequence positions (−242 to −230) in Group D (Fig 2). The significance of these changes remains to be investigated.
The high degree of homology between Group D, the Asian isolates, CAO, and clone1510 was restricted to changes in the promoter region and loss of the Xho-I restriction site. In contrast, only 48% of the mutations found in the LMP-1 gene of Group D isolates were identical to CAO11 mutations (Fig 1). C151013 and Raji11 (Fig 1) showed 53% and 21% homology, respectively. Most importantly, the Group D isolates do not contain the 30-bp deletion shown to increased the tumorigenicity of LMP-1 in vitro.25 This clearly shows that the CAO-LMP-1–associated Xho I restriction polymorphism at position 169425 and the C-terminal 30-bp deletion occur independently of each other in wild-type isolates. Similar findings have been reported for tumor-derived isolates by Miller et al.16 Thus, Western EBV isolates that have only been investigated for the 30-bp deletion and Asian NPC viruses only characterized for Xho I polymorphism11,13-15,18-20 may be similar to our Group C and Group D isolates, respectively, rather than to CAO as has previously been assumed.11 This suggests that loss of the Xho I restriction site should not be used as a marker for an LMP-1 protein with increased oncogenicity, but rather as a marker of a weaker promoter. The C-terminal effector region of the LMP-1 protein (designated CTAR-2 by Huen et al), shown to be important for LMP-1 induced NF-kB activation and possibly overlapping the 30-bp deletion,27 contains three positively charged amino acids (all histidines). Interestingly, two of these histidines (352 and 358) have been replaced by asparagine and proline, respectively, in all four Group D isolates, whereas two of the isolates have lost all three histidines (352, 358, 377), the last one substituted with asparagine. How these changes affect the function of LMP-1 is not known.
The number of 33-bp repeats in our isolates varied from 3 to 7 (Table 1 and Fig 1) but the configuration of the repeat region did not by itself define separate groups. Similar findings have been reported previously.16 However, since only Group A isolates contained repeats identical to B95.8, identification of a B95.8-like repeat region indicates that no (or only few) single base mutations can be expected in the LMP-1 gene.
Another interesting amino acid change is the substitution of methionine129 with isoleucine found in Groups B, C, and D, this site only being conserved in the seven isolates with an LMP-1 gene similar to the B.95.8 strain (Group A ) (Fig 3). Methionine 129 has been suggested as the translation initiation site for the so-called truncated or lytic form of LMP-1.31 This may partly explain why previous reports have found that TPA stimulation only induces truncated LMP-1 in B95.8 but not in other virus isolates.32
Miller et al16 have previously reported partial LMP-1 sequence for a range of isolates from various Asian, Middle-Eastern, North American, and Eskimo EBV-associated tumors. Wild-type isolates from individuals without malignant disease were not studied. Direct correlation of their data with our proposed virus groups is difficult as (with the exception of their isolate C15) only parts of the LMP-1 N- and C-terminal regions were fully sequenced. These authors also found that the LMP-1 gene was generally highly conserved, but they described a somewhat wider spread of sequence variations in their tumor-derived isolates (especially in the C-terminus) with 41 of the 71 (57%) single base mutations identified being detected in only a single isolate.17 In the same region, our results show fewer single base mutations overall (31) of which only 3 (10%) were confined to a single isolate. However, if only mutations detected in more than one isolate are compared, there is a quite high degree of concordance between our isolates and those reported by Miller et al. Thus, comparing only Asian, Middle-Eastern, and North American NPC isolates, 15 of the 16 N-terminal and all 14 of the C-terminal single base mutations they detected are found in our Group A through D isolates. Similarly, their isolates with the 30-bp deletion (with one exception) are intermediate between our Group C and CAO whereas those without the 30-bp deletion are similar to our Group B viruses. One isolate from a PTL (designated HE) is identical to our Group D.16 The Eskimo and BL isolates described by Miller et al show a lower degree of homology to our isolates. These authors also describe 14 single base mutations in the LMP-1 N-terminus in all isolates with the Xho I polymorphism. Although our Group D viruses contain 13 of these 14 mutations, 10 are also seen in the Group C indicating that most are not specific for the loss of the Xho I site.
Knecht et al33 have also investigated the frequency of mutations in the C-terminus of the LMP-1 gene in various EBV-positive lymphoproliferative disorders. They described a number of isolates that differed from B95.8, all of which can be allocated to one of our Groups A through C. No isolates similar to Group D were found. Knecht et al33 suggested that clustered amino acid substitutions at positions 322, 334, 338, and 366 were more frequent in isolates from patients with lymphoproliferative disorders than in individuals without EBV-associated disease. In a previous study, we found similar amino acid substitutions at these four positions in isolates from 39% of Danish HD and PTLs.17 In the present study, identical substitutions were identified in all our Group B and Group C isolates, representing some 38% of EBV isolates from individuals without EBV-related disease. Thus, while some of these sites appear to represent mutational hot spots, as previously suggested,17,33 our data does not support the suggestion by Knecht et al33 that they are more frequent in isolates from EBV-positive lymphoproliferative disorders.
Supported by the Danish Cancer Society, Copenhagen, Denmark, Grant No. 95 120 01 and 95 100 16.
Address reprint requests to Kristian Sandvej, MD, Laboratory of Immunopathology, University Institute of Pathology, Aarhus Kommunehospital, Finsensgade 12, DK-8000 Aarhus C, Denmark.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal