Abstract
Interleukin-6 (IL-6) is a growth factor for multiple myeloma (MM) cells and can inhibit MM cell apoptosis. Our recent studies show that IL-6 facilitates MM cell growth via phosphorylation of retinoblastoma protein (pRB); however, the effects of IL-6 on those cyclins, cyclin-dependent kinases (CDKs), and CDK inhibitors (CDIs) that are known to regulate phosphorylation of pRB have not been defined in MM cells. In the present report, we cultured MM cell lines and patient cells with IL-6 and/or dexamethasone (Dex) and characterized changes in cell cycle; expression and association of cyclins, CDKs, and CDIs; and phosphorylation of pRB. Dex induced G1 growth arrest in MM cells, whereas IL-6 facilitated G1 to S phase transition; moreover, the effect of Dex was blocked by IL-6. p21WAF1 (p21) protein was constitutively expressed in the majority of MM cells independent of the status of p53. Its expression was upregulated by Dex and downregulated by IL-6; again, IL-6 inhibited the increase in p21 triggered by Dex. These alterations in p21 expression in MM cells were associated with changes in p21 binding to CDK2, CDK4, and CDK6; CDK2, CDK4, and CDK6 kinase activities; and phosphorylation of pRB. In contrast, expression of G1 cell cycle regulatory proteins, including p27KIP1, cyclin D2, and cyclin E, was not altered in MM cells cultured with Dex and/or IL-6. Finally, interferon-γ (IFN-γ) also induced G1 growth arrest and upregulated p21 protein expression; as with Dex, affects of IFN-γ were inhibited by IL-6. Our results therefore show that changes in cell cycle distribution in MM cells triggered by Dex, IL-6, and IFN-γ correlate with changes in p21 protein expression and implicate p21 in the coupling of Dex-, IL-6–, and IFN-γ–related signals to G1 cell cycle regulation in MM cells.
GROWTH AND differentiation of normal and malignant cells can be regulated by a variety of external stimuli, ie, cytokines or cell-cell and cell-extracellular matrix interactions. The same growth factor may have differential effects on normal and malignant cells. For example, interleukin-6 (IL-6) is a differentiation factor for normal B cells but is an autocrine and paracrine growth factor, as well as an antiapoptotic factor, for multiple myeloma (MM) cells.1-11 IL-1β, tumor necrosis factor-α, transforming growth factor β, interferons (IFNs), CD40 ligand, dexamethasone (Dex), retinoic acid, and adhesion to bone marrow (BM) stromal cells also effect MM cell growth and survival, and many of these stimuli either modulate production of IL-6 or expression of IL-6 receptor (IL-6R) on tumor cells.12-22 For example, Dex inhibits proliferation and induces apoptosis of MM cells; it can induce apoptosis of MM cells growing in an IL-6–mediated autocrine mechanism by both blocking IL-6 production and by inhibiting IL-6R expression on tumor cells.9 Dex can also induce growth arrest and apoptosis in MM cells that grow independently of IL-6; however, even in this setting, IL-6 can inhibit Dex-related affects.23 Finally, IFN represents another example of therapy for MM that may, at least in part, work by downregulating IL-6R expression on MM cells.14-17 Delineation of the molecular targets whereby these external stimuli regulate MM cell growth and survival might not only enhance our understanding of disease pathogenesis, but also both elucidate the mechanisms of current treatments and suggest novel therapies.
IL-6–related signal transduction pathways have been defined in both normal and malignant cells.24 Specifically, IL-6 binds to its α chain receptor (gp80) and induces homodimerization of gp130 and activation of the intracytoplasmic Janus family of tyrosine kinases (JAKs), with downstream signaling via the signal transducers and activators of transcription (STAT) or Ras-dependent mitogen-activated protein kinase (MAPK) cascades.24 Moreover, we have recently shown that IL-6 facilitates MM cell growth via phosphorylation of retinoblastoma protein (pRB).25 To date, a variety of cyclins, cyclin-dependent kinases (CDKs), and CDK inhibitors (CDIs) are known to regulate phosphorylation of pRB26; however, which of these proteins are associated with cell cycle control in MM cells is not yet defined.
In the present report, we investigated the effects of IL-6, Dex, or both on cyclins, CDKs, and CDIs affecting phosphorylation of pRB in both MM-derived cell lines and patient MM cells. Dex induced G1 growth arrest in MM cells, an effect that was inhibited by IL-6. Dex upregulated and IL-6 downregulated p21WAF1 (p21) expression in MM cells in a time- and dose-dependent fashion, and IL-6 prevented this p21 upregulation. Changes in p21 expression in MM cells were associated with changes in its binding to CDK2, CDK4, and CDK6; CDK2, CDK4, and CDK6 kinase activities; phosphorylation of pRB; and cell cycle progression. As with Dex, IFN-γ induced G1 growth arrest and decreased p21 expression in MM cells; IL-6 similarly inhibited these IFN-γ–related effects. Our results therefore show that changes in cell cycle distribution triggered by Dex, IL-6, and IFN-γ are correlated with changes in p21 protein expression in MM cells.
MATERIALS AND METHODS
Preparation of cells.Mononuclear cells (MCs) were isolated from five patients with MM by Ficoll-Hypaque (FH) density gradient centrifugation and then incubated with HB7 (anti-CD38) monoclonal antibody (MoAb)-biotin-streptavidin and 2H4 (anti-CD45RA) MoAb-fluorescein isothiocyanate on ice. Tumor cells (>95% CD38+CD45RA−) were isolated using an Epics C Cell Sorter (Coulter Electronics, Hialeah, FL), washed, and resuspended in RPMI-1640 media (Sigma Chemical Co, St Louis, MO) containing 10% fetal bovine serum (FBS), L-glutamine (L-glu; GIBCO, Grand Island, NY), 100 U/mL penicillin (pen), and 100 μg/mL streptomycin (strep; GIBCO). The RPMI-8226, ARH-77, U-266, and HS Sultan human MM-derived cell lines4 were obtained from American Type Culture Collection (Rockville, MD). The IL-6–responsive S6B45 MM-derived cell line27 was provided by Dr H. Suzuki (Osaka University, Osaka, Japan). We established the JKB cell line from a patient with pre-B acute lymphoblastic leukemia (ALL).28 These cell lines were cultured in RPMI-1640 media (Sigma) with 10% FBS, L-glu, and pen/strep. The OCI-My5 MM cell line5 was kindly provided by Dr H.A. Messner (Ontario Cancer Institute, Toronto, Ontario, Canada) and cultured in Iscove's media (Sigma) with 10% FBS, L-glu, and pen/strep.
BM specimens were obtained from healthy donors and MCs separated by FH density sedimentation. Single-cell suspensions from normal spleen were prepared by extrusion through sterile stainless steel mesh. To obtain normal human B cells (>90% CD20+), splenic MCs were first isolated by FH density sedimentation. Monocytes and T cells were next depleted by double adherence to plastic and by rosetting with sheep red blood cells, respectively, as previously described.13
Enhancement of wild-type p53 protein expression by γ-irradiation.To enhance wild-type p53 protein expression, cells were irradiated (3.0 Gy) and lysed 2 hours later. Increased wild-type p53 protein expression was confirmed by immunoprecipitation and Western blotting, as described below.
Cell cycle analysis.MM-derived cell lines (2 × 105/mL) and patient MM cells (1 × 106/mL) were cultured with media alone, Dex, IL-6, IFN-γ, and Dex or IFN-γ + IL-6. Cells were harvested at intervals, washed with phosphate-buffered saline, stained with propidium iodide (PI), and analyzed using fluorescence-activated cell sorting (FACS).27 Specifically, cells were collected and resuspended in 0.5 mL of 3.4 mmol/L sodium citrate, 10 mmol/L NaCl, 0.1% NP-40, and 50 μg/mL PI to stain nuclear DNA. Fluorescence intensity was measured using the Coulter Epics 753 Cell Sorter (Coulter Immunology, Hialeah, FL). For each sample, 10,000 cells were analyzed and results were processed using program M Cycle software (Coulter).
Immunoprecipitation and Western immunoblotting.Immunoprecipitation and Western immunoblotting were performed as previously reported.26 For immunoprecipitation, cells (1 × 107 cells/sample) were cultured in 10% FBS-RPMI-1640 media or 10% FBS-Iscove's media, washed three times with phosphate-buffered saline, and lysed for 30 minutes at 4°C in buffer: 50 mmol/L Tris-HCl (pH 8.0), 150 mmol/L NaCl, 0.1% Nonidet P-40, 1 mmol/L EDTA, 5 μg/mL phenylmethyl sulfonyl fluoride, 1 mmol/L Na3VO4 , 2 μg/mL aprotinin, 2 μg/mL leupeptin, and 50 mmol/L NaF. Antibodies against cell cycle regulatory proteins were added and incubated for 16 hours at 4°C. Proteins were collected using protein G sepharose (PGS). Aliquots of each lysate were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Protein was transferred onto nitrocellulose membranes, and nonspecific binding was blocked by incubation with 5% skim milk. The membranes were probed with antibodies, incubated with antimouse or antirabbit Ig antibodies conjugated with horseradish peroxidase (Amersham, Arlington Heights, IL), and detected using the enhanced chemiluminescence system (Amersham).
Immune complex kinase assays.Immune complex kinase assays were performed using the method of Matsushime et al,29 with modification. Briefly, cells were suspended at 1 × 107/mL in the lysis buffer described above and centrifuged at 10,000g for 5 minutes, and the supernatants were precipitated for 16 hours at 4°C with PGS plus rabbit anti-CDK2, anti-CDK4, or anti-CDK6 polyclonal Abs. Immunoprecipitated proteins on PGS were washed three times with 1 mL of lysis buffer and twice with 50 mmol/L HEPES (pH 7.5) containing 1 mmol/L dithiothreitol and suspended in 30 μL of kinase buffer (50 mmol/L HEPES, 10 mmol/L MgCl2 , 1 mmol/L dithiothreitol) containing substrate and 2.5 mmol/L EGTA, 10 mmol/L β-glycerophosphate, 0.1 mmol/L sodium orthovanadate, 1 mmol/L NaF, 20 μmol/L ATP, and 10 μCi of γ-32P-ATP (NEN Dupont, Boston, MA). For CDK2 kinase assays, 1 μg of histone H1 (Boehringer Mannheim, Indianapolis, IN) was the substrate; for CDK4 and CDK6 kinase assays, 1 μg of soluble glutathione S-transferase (GST)-RB fusion protein (Santa Cruz Biotechnology, Santa Cruz, CA) was the substrate. After incubation for 30 minutes at 30°C with occasional mixing, the samples were boiled in polyacrylamide gel sample buffer and separated by SDS electrophoresis. Phosphorylated proteins were visualized by autoradiography of the dried slab gels. The intensity was quantitated with densitometry compared with media alone.
Reagents.IL-6 (Kirin-Brewery Co Ltd, Tokyo, Japan) and/or Dex (Sigma) or IFN-γ (Endogen, Cambridge, MA) were added to cultures of patient MM cells and MM-derived cell lines; the effects on cell cycle distribution and kinetics of cell cycle protein expression and association were assayed. Immunoprecipitation and Western immunoblotting were performed using mouse anti-p27 KIP1 (p27) MoAb, anti-p21 MoAb, anti-pRB MoAb, or anti-actin MoAb (Oncogene Science, Uniondale, NY); with rabbit anti-p21, anti-p27, anti-CDK2, anti-CDK4 (carboxy terminus), anti-CDK-6 (carboxy terminus), anti-cyclin D2, and anti-cyclin E (Santa Cruz Biotechnology) polyclonal Abs; and with mouse anti-cyclin E MoAb (Santa Cruz Biotechnology).
To detect wild-type and/or mutated p53 protein, cell lysates were immunoprecipitated with Ab-5 anti-p53 MoAb, which recognizes wild-type p53, or with Ab-3 anti-p53 MoAb, which recognizes mutated type p53, and then immunoblotted with Ab-6 anti-p53 MoAb (Oncogene Science) or DO-1 horseradish peroxidase-conjugated anti-p53 MoAb recognizing pantropic p53 (Santa Cruz Biotechnology).
RESULTS
Expression of wild-type and mutated p53, as well as p21 and p27, in MM cell lines and patient MM cells.Expression of wild-type and mutated p53, as well as p21 and p27, in MM cells is shown in Table 1. Immunoprecipitation studies showed that all six MM cell lines expressed mutated p53 protein. Specifically, RPMI-8226, U-266, and HS Sultan expressed mutated p53 protein, with loss of heterozygosity, because wild-type p53 remained undetectable even after γ-irradiation. p53 protein expression in these MM cell lines was consistent with a previous report of p53 gene using polymerase chain reaction single-stranded conformation polymorphism analysis.30 In contrast, three MM cell lines (OCI-My5, S6B45, and ARH-77) expressed both wild-type and mutated p53 proteins. The JKB ALL cell line showed increased wild-type p53 protein expression induced by irradiation, without detectable mutated p53.
Expression of Wild-Type and Mutated p53 as Well as p21 and p27 in MM Cell Lines and MM Cells of Patients
| Cells . | Wt p53* . | Mt p53† . | p21‡ . | p27‡ . |
|---|---|---|---|---|
| OCI-My5 | + | + | + | ++ |
| RPMI-8226 | − | +++ | + | ++ |
| U-266 | − | +++ | + | + |
| HSS | − | +++ | + | +++ |
| S6B45 | + | + | ++ | +++ |
| ARH-77 | + | +++ | − | − |
| JKB | + | − | ++ | + |
| Patient no. 1 | + | + | + | ++ |
| Patient no. 2 | − | + | + | ++ |
| Patient no. 3 | + | − | + | + |
| Patient no. 4 | + | − | + | ++ |
| Patient no. 5 | + | − | + | ++ |
| Cells . | Wt p53* . | Mt p53† . | p21‡ . | p27‡ . |
|---|---|---|---|---|
| OCI-My5 | + | + | + | ++ |
| RPMI-8226 | − | +++ | + | ++ |
| U-266 | − | +++ | + | + |
| HSS | − | +++ | + | +++ |
| S6B45 | + | + | ++ | +++ |
| ARH-77 | + | +++ | − | − |
| JKB | + | − | ++ | + |
| Patient no. 1 | + | + | + | ++ |
| Patient no. 2 | − | + | + | ++ |
| Patient no. 3 | + | − | + | + |
| Patient no. 4 | + | − | + | ++ |
| Patient no. 5 | + | − | + | ++ |
Protein expression levels were compared in the same number of cells (−, undetectable; +, weakly expressed; ++, moderately expressed; +++, highly expressed).
Cells were irradiated at 3 Gy.
Expression was not altered by irradiation.
Cells were not irradiated.
Two of five patients' MM cells expressed mutated p53 protein, which was associated with loss of heterozygosity in a single case. The remaining three patient MM cells expressed wild-type p53 protein, induced by irradiation, without detectable mutated p53 protein. Except for ARH-77 cells, p21 and p27 proteins were expressed in all MM cell lines and patient MM samples, including cells expressing wild-type and/or mutant p53.
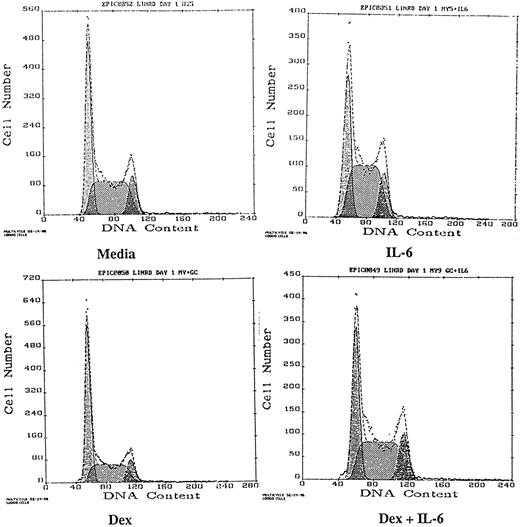
Effects of Dex and/or IL-6 on cell cycle distribution and p21 protein expression in MM cell lines and patient MM cells.MM-derived cell lines and patient MM cells were cultured with media alone, IL-6, Dex, or both IL-6 and Dex for 16 hours, and cell cycle distribution was analyzed by PI staining and FACS analysis (Table 2). The effect of Dex and/or IL-6 on cell cycle distribution of OCI-My5 MM cells is also shown in Fig 1. IL-6 stimulated a 5% to 15% shift from G1 to S/G2M in OCI-My5 and U-266 cells, as well as in the cells of MM patients no. 1, 3, and 5. Dex alone (0.1 to 1.0 μmol/L) induced G1 growth arrest in all MM cell lines and in patient MM cells, as evidenced by a 10% to 25% increase in G1 and a decrease in S relative to media alone cultures, except in ARH-77 MM cells, which were resistant to Dex. Finally, culture with both Dex and IL-6 showed that IL-6 blocked the G1 growth arrest induced by Dex in OCI-My5 and S6B45 cells, as well as in the cells of MM patients no. 1, 3, and 5. MM cell lines and MM patient cells were defined as IL-6 responsive if IL-6 shifted the fraction of MM cells from G1 to S by ≥10% and/or inhibited Dex-induced G1 growth arrest by ≥10%. Viability of all MM cell lines and patient MM cells remained high (>95%) during the 16 hours of culture with media, IL-6, Dex, or both.
Effects of IL-6 and/or Dex on Cell Cycle Distribution on Multiple Myeloma Cells
| Cells* . | % Cells in G1 Phase . | |||||||
|---|---|---|---|---|---|---|---|---|
| . | Media . | IL-6 . | Dex (μmol/L) . | Dex + IL-6 . | . | . | ||
| . | . | . | 0.01 . | 0.10 . | 1.00 . | . | . | . |
| OCI-My5 | 41.5 | 31.8 | 42.3 | 50.2 | 62.5 | 34.6 | ||
| RPMI-8226 | 37.5 | 36.8 | 37.5 | 40.3 | 58.9 | 54.3 | ||
| U-266 | 67.5 | 57.7 | 66.2 | 84.6 | 92.3 | 90.4 | ||
| HSS | 44.5 | 43.4 | 45.3 | 51.5 | 63.8 | 62.7 | ||
| S6B45 | 62.6 | 59.2 | 64 | 83.2 | 89.4 | 66.3 | ||
| ARH-77 | 55 | 57.3 | 54.2 | 55.6 | 57.1 | 53.3 | ||
| JKB | 79.3 | 80.2 | 78.3 | 85.9 | 91.8 | 90.9 | ||
| Patient no. 1 | 81.5 | 74.3 | 85.9 | 93.3 | 92.5 | 77.6 | ||
| Patient no. 2 | 85.1 | 84.6 | 90.2 | 93.6 | 95.3 | 90.6 | ||
| Patient no. 3 | 88.7 | 74.2 | 89.9 | 94.2 | 96.6 | 90.6 | ||
| Patient no. 4 | 87 | 82.4 | 91 | 92.9 | 93.3 | 91 | ||
| Patient no. 5 | 78.2 | 71.3 | 90.5 | 93.7 | 94.6 | 82.3 | ||
| Cells* . | % Cells in G1 Phase . | |||||||
|---|---|---|---|---|---|---|---|---|
| . | Media . | IL-6 . | Dex (μmol/L) . | Dex + IL-6 . | . | . | ||
| . | . | . | 0.01 . | 0.10 . | 1.00 . | . | . | . |
| OCI-My5 | 41.5 | 31.8 | 42.3 | 50.2 | 62.5 | 34.6 | ||
| RPMI-8226 | 37.5 | 36.8 | 37.5 | 40.3 | 58.9 | 54.3 | ||
| U-266 | 67.5 | 57.7 | 66.2 | 84.6 | 92.3 | 90.4 | ||
| HSS | 44.5 | 43.4 | 45.3 | 51.5 | 63.8 | 62.7 | ||
| S6B45 | 62.6 | 59.2 | 64 | 83.2 | 89.4 | 66.3 | ||
| ARH-77 | 55 | 57.3 | 54.2 | 55.6 | 57.1 | 53.3 | ||
| JKB | 79.3 | 80.2 | 78.3 | 85.9 | 91.8 | 90.9 | ||
| Patient no. 1 | 81.5 | 74.3 | 85.9 | 93.3 | 92.5 | 77.6 | ||
| Patient no. 2 | 85.1 | 84.6 | 90.2 | 93.6 | 95.3 | 90.6 | ||
| Patient no. 3 | 88.7 | 74.2 | 89.9 | 94.2 | 96.6 | 90.6 | ||
| Patient no. 4 | 87 | 82.4 | 91 | 92.9 | 93.3 | 91 | ||
| Patient no. 5 | 78.2 | 71.3 | 90.5 | 93.7 | 94.6 | 82.3 | ||
Cell cycle distribution was examined by PI staining followed by FACS analysis.
Cell lines (2 × 105) and patients' MM cells (1 × 106) were cultured for 16 hours with media alone, IL-6 (50 ng/mL), Dex, or IL-6 (50 ng/mL) + Dex (1 μmol/L).
Effects of Dex and/or IL-6 on cell cycle distribution of OCI-My5 MM cells. OCI-My5 cells (2 × 105/mL) were cultured with media alone, IL-6 (50 ng/mL), Dex (1 μmol/L), or both IL-6 (50 ng/mL) and Dex (1 μmol/L). Cell cycle distribution was analyzed by PI staining and FACS 16 hours later.
Effects of Dex and/or IL-6 on cell cycle distribution of OCI-My5 MM cells. OCI-My5 cells (2 × 105/mL) were cultured with media alone, IL-6 (50 ng/mL), Dex (1 μmol/L), or both IL-6 (50 ng/mL) and Dex (1 μmol/L). Cell cycle distribution was analyzed by PI staining and FACS 16 hours later.
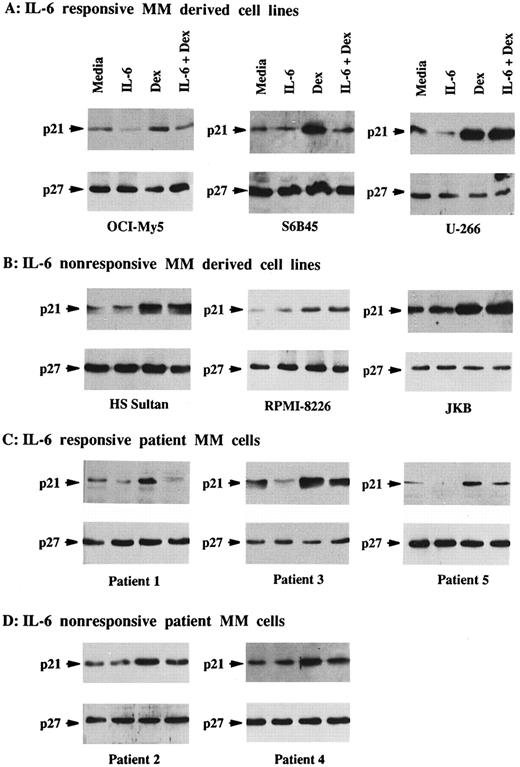
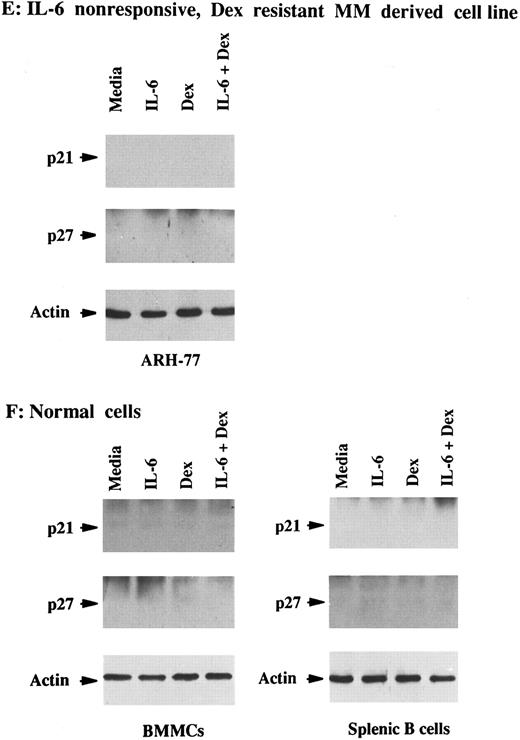
Total lysates were next made from five MM cell lines and five MM patient cells cultured under similar conditions. p21 was constitutively expressed in Dex-sensitive MM cell lines that were both responsive to IL-6 (OCI-My5, U-266, and S6B45) and nonresponsive to IL-6 (HS Sultan and RPMI-8226), as well as in IL-6–nonresponsive JKB ALL cells (Fig 2A and B). It was similarly constitutively expressed in both IL-6–responsive (patients no. 1, 3, and 5) and IL-6–nonresponsive (patients no. 2 and 3) patient MM cells (Fig 2C and D). In contrast, p21 was not detectable in ARH-77 MM cells (Fig 2E), normal B cells, or normal BMMCs (Fig 2F ). IL-6 downregulated p21 protein expression in OCI-My5 and U-266 MM cells as well as in the cells of MM patients no. 1, 3, and 5; however, it had no affect on p21 expression in RPMI-8226, HS Sultan, and S6B45 MM cells as well as the cells of MM patients no. 2 and 4. Dex upregulated p21 expression both in patient MM cells and in all MM-derived cell lines, except for ARH-77 cells. Cultures with both Dex and IL-6 showed that the increases in p21 expression triggered by Dex were blocked by IL-6 in OCI-My5 and S6B45 MM cells and in the MM cells of patients no. 1, 3, and 5; however, IL-6 did not inhibit upregulation of p21 induced by Dex in RPMI-8226, HS Sultan, and U-266 MM cells and in the MM cells of patients no. 2 and 4. p21 remained undetectable in ARH-77 MM cells, normal B cells, and normal BMMCs cultured with IL-6, Dex, or both. Finally, p27 protein expression was not altered by Dex and/or IL-6 in these same cell lysates from either MM cell lines or patient MM cells.
Effects of Dex and/or IL-6 on p21 protein expression in MM cell lines and patient MM cells. Cells (2 × 105/mL) from IL-6–responsive MM-derived cell lines (OCI-My5, S6B45, and U-266; A) and IL-6–nonresponsive MM cell lines (HS Sultan and RPMI-8226) and JKB ALL cell line (B); 1 × 106/mL IL-6–responsive patient MM cells (patients no. 1, 3, and 5; C) and IL-6–nonresponsive patient MM cells (patients no. 2 and 4; D); 2 × 105/mL IL-6–nonresponsive Dex-resistant ARH-77 MM cells (E); and 1 × 106/mL normal BMMCs and splenic B cells (F ) were cultured with 10% FBS media alone, IL-6 (50 ng/mL), Dex (1 μmol/L), or Dex (1 μmol/L) and IL-6 (50 ng/mL). Total cell lysates were prepared 16 hours later and immunoprecipitated with anti-p21 or anti-p27 polyclonal Abs, followed by Western immunoblotting with anti-p21 or anti-p27 MoAbs. Immunoprecipitation and immunoblotting with anti-actin MoAb confirmed equal protein loading in cells that lacked detectable p21 and p27: ARH-77 (E) and normal BMMCs and splenic B cells (F ).
Effects of Dex and/or IL-6 on p21 protein expression in MM cell lines and patient MM cells. Cells (2 × 105/mL) from IL-6–responsive MM-derived cell lines (OCI-My5, S6B45, and U-266; A) and IL-6–nonresponsive MM cell lines (HS Sultan and RPMI-8226) and JKB ALL cell line (B); 1 × 106/mL IL-6–responsive patient MM cells (patients no. 1, 3, and 5; C) and IL-6–nonresponsive patient MM cells (patients no. 2 and 4; D); 2 × 105/mL IL-6–nonresponsive Dex-resistant ARH-77 MM cells (E); and 1 × 106/mL normal BMMCs and splenic B cells (F ) were cultured with 10% FBS media alone, IL-6 (50 ng/mL), Dex (1 μmol/L), or Dex (1 μmol/L) and IL-6 (50 ng/mL). Total cell lysates were prepared 16 hours later and immunoprecipitated with anti-p21 or anti-p27 polyclonal Abs, followed by Western immunoblotting with anti-p21 or anti-p27 MoAbs. Immunoprecipitation and immunoblotting with anti-actin MoAb confirmed equal protein loading in cells that lacked detectable p21 and p27: ARH-77 (E) and normal BMMCs and splenic B cells (F ).
Time- and dose-dependent effects of Dex and/or IL-6 on cell cycle distribution and p21 protein expression in OCI-My5 MM cells.The effects of culture with IL-6, Dex, or both on the cell cycle distribution of OCI-My5 MM cells during 48 hours of culture was examined using PI staining followed by FACS analysis (Table 3). Cultures with media alone showed no significant changes in cell cycle distribution. Dex induced significant increases in percentage of cells in G1 at 16 hours, with corresponding decreases in S; in contrast, culture with IL-6 decreased the fraction of cells in G1, with associated increases in S, at this time. Finally, in the presence of both Dex and IL-6, the percentage of cells in G1 decreased and S increased to a lesser extent, but in a similar pattern, to that observed with IL-6 alone. In these experiments, viability of OCI-My5 MM cells remained high (>95%) during 48 hours of culture with media, Dex, IL-6, or both Dex + IL-6.
Time-Dependent Effects of Dex and/or IL-6 on Cell Cycle Distribution of OCI-My5 Cells
| . | Time (h) . | |||||||
|---|---|---|---|---|---|---|---|---|
| . | 0 . | 2 . | 4 . | 8 . | 16 . | 24 . | 36 . | 48 . |
| Media | 40.5 | 40.7 | 41 | 41.3 | 41.5 | 42.7 | 43 | 43.1 |
| IL-6 (50 ng/mL) | 40.5 | 40.8 | 39.2 | 38.8 | 31.8 | 31 | 30.2 | 31.4 |
| Dex (1 μmol/L) | 40.5 | 40.7 | 45.7 | 55.7 | 62.5 | 62.6 | 62.2 | 62.8 |
| IL-6 (50 ng/mL) + Dex (1 μmol/L) | 40.5 | 41.6 | 40.9 | 39.3 | 34.6 | 33.7 | 33.5 | 32.3 |
| . | Time (h) . | |||||||
|---|---|---|---|---|---|---|---|---|
| . | 0 . | 2 . | 4 . | 8 . | 16 . | 24 . | 36 . | 48 . |
| Media | 40.5 | 40.7 | 41 | 41.3 | 41.5 | 42.7 | 43 | 43.1 |
| IL-6 (50 ng/mL) | 40.5 | 40.8 | 39.2 | 38.8 | 31.8 | 31 | 30.2 | 31.4 |
| Dex (1 μmol/L) | 40.5 | 40.7 | 45.7 | 55.7 | 62.5 | 62.6 | 62.2 | 62.8 |
| IL-6 (50 ng/mL) + Dex (1 μmol/L) | 40.5 | 41.6 | 40.9 | 39.3 | 34.6 | 33.7 | 33.5 | 32.3 |
OCI-My5 cells (5 × 105) were cultured with 10% FBS Iscove's media alone, IL-6, Dex, or IL-6 + Dex. Cells (1 × 106) were harvested at intervals, washed, stained with PI, and analyzed using FACS. Values are the percentage of cells in G1 phase.
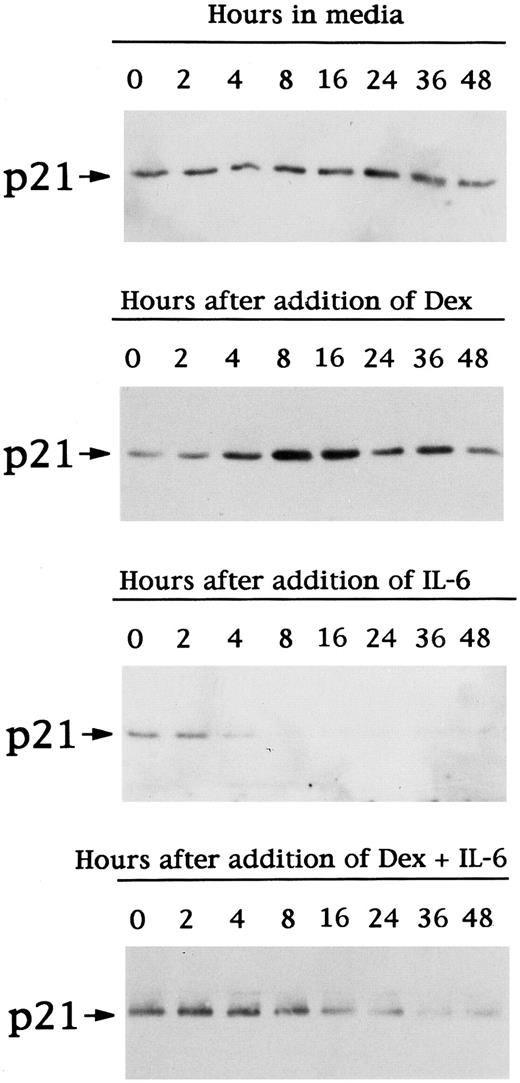
To assay for changes in p21 expression associated with these changes in cell cycle distribution, we next examined p21 protein expression in cell lysates from OCI-My5 MM cells cultured under the same conditions (Fig 3). p21 protein was constitutively expressed and unchanged throughout 48 hours of culture in media. Dex triggered peak increases in p21 protein expresssion at 8 to 16 hours, which returned to baseline at 48 hours. In the presence of IL-6, p21 protein expression decreased with time. Finally, culture with both Dex and IL-6 also decreased p21 expression, but to a lesser extent than noted in lysates from cells cultured with IL-6 alone.
Time-dependent effects of Dex and/or IL-6 on expression of p21 protein in OCI-My5 MM cells. Total lysates from OCI-My5 MM cells were prepared before (time 0) and after 2, 4, 8, 16, 24, 36, and 48 hours of culture with 10% FBS-Iscove's media alone, Dex (1 μmol/L), IL-6 (50 ng/mL), or Dex (1 μmol/L) + IL-6 (50 ng/mL). Cell lysates were immunoprecipitated with anti-p21 polyclonal Ab, followed by Western immunoblotting with anti-p21 MoAb.
Time-dependent effects of Dex and/or IL-6 on expression of p21 protein in OCI-My5 MM cells. Total lysates from OCI-My5 MM cells were prepared before (time 0) and after 2, 4, 8, 16, 24, 36, and 48 hours of culture with 10% FBS-Iscove's media alone, Dex (1 μmol/L), IL-6 (50 ng/mL), or Dex (1 μmol/L) + IL-6 (50 ng/mL). Cell lysates were immunoprecipitated with anti-p21 polyclonal Ab, followed by Western immunoblotting with anti-p21 MoAb.
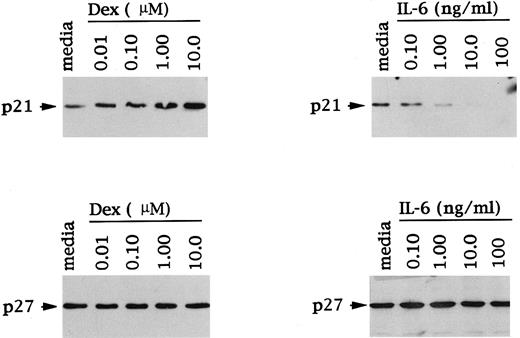
The dose response of Dex and IL-6 effects on p21 expression was next examined, again using OCI-My5 MM cells. As can be seen in Fig 4, Dex (0.01 to 10 μmol/L) triggered a dose-dependent increase in p21 protein expression. Moreover, IL-6–related decreases in p21 protein expression were also dependent on IL-6 dose. In contrast, levels of p27 protein expression in these same cell lysates were unchanged in the presence of Dex and/or IL-6 and confirmed equal protein loading.
Dose-dependent effects of Dex and IL-6 on p21 and p27 expression in MM cells. Total cell lysates were prepared from OCI-My5 MM cells after 16 hours of culture with Dex (0.01 to 10 μmol/L) or IL-6 (0.10 to 100 ng/mL). Lysates were immunoprecipitated with anti-p21 or anti-p27 polyclonal Abs, followed by Western immunoblotting with anti-p21 or anti-p27 MoAbs.
Dose-dependent effects of Dex and IL-6 on p21 and p27 expression in MM cells. Total cell lysates were prepared from OCI-My5 MM cells after 16 hours of culture with Dex (0.01 to 10 μmol/L) or IL-6 (0.10 to 100 ng/mL). Lysates were immunoprecipitated with anti-p21 or anti-p27 polyclonal Abs, followed by Western immunoblotting with anti-p21 or anti-p27 MoAbs.
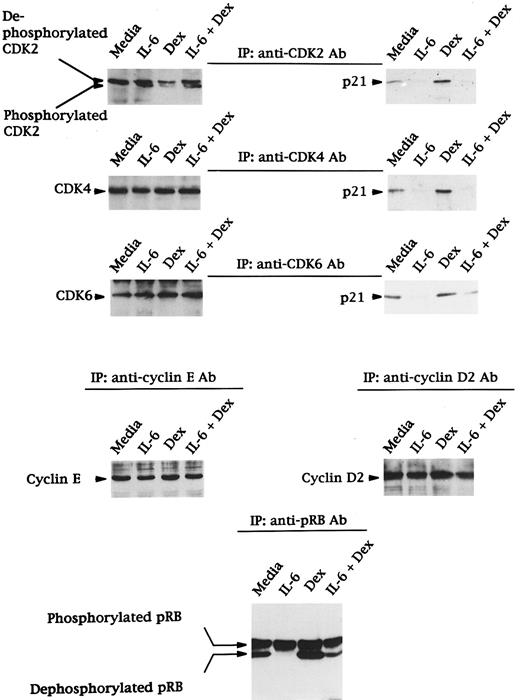
Effects of Dex and/or IL-6 on expression of CDK2, CDK4, and CDK6; binding of p21 to CDK2, CDK4, and CDK6; expression of cyclin E and D2; and phosphorylation of RB in OCI-My5 cells.The observations that Dex induced G1 growth arrest and that IL-6 facilitated entry from G1 to S phase in OCI-My5 MM cells, with corresponding changes in p21 protein expression, prompted us to next examine expression of other proteins (CDK2, CDK4, CDK6, cyclin D2, cyclin E, and pRB) known to regulate G1/S transition, as well as the binding of CDK2, CDK4, and CDK6 to p21 (Fig 5). CDK2 was constitutively expressed in its dephosphorylated form in media cultures; phosphorylation was induced by IL-6 and downregulated by Dex. Moreover, IL-6 inhibited the decrease in phosphorylation of CDK2 triggered by Dex. CDK4 and CDK6 were also constitutively expressed in media cultures, and their expression was unchanged in the presence of IL-6 and/or Dex.
Effects of Dex and/or IL-6 on expression of CDK2, CDK4, and CDK6; binding of p21 to CDK2, CDK4, and CDK6; expression of cyclin E and D2; and phosphorylation of RB in OCI-My5 MM cells. Total cells lysates were prepared from OCI-My5 MM cells cultured with media alone, IL-6 (50 ng/mL), Dex (1 μmol/L), or IL-6 (50 ng/mL) + Dex (1 μmol/L) for 16 hours. Lysates were immunoprecipitated with anti-CDK2, anti-CDK4, or anti-CDK6 polyclonal antibodies followed by Western immunoblotting with either the same antibodies or anti-p21 MoAb. Expression of cyclin E, cyclin D2, and pRB were analyzed by immunoprecipitation and Western immunoblotting using the same cell lysates.
Effects of Dex and/or IL-6 on expression of CDK2, CDK4, and CDK6; binding of p21 to CDK2, CDK4, and CDK6; expression of cyclin E and D2; and phosphorylation of RB in OCI-My5 MM cells. Total cells lysates were prepared from OCI-My5 MM cells cultured with media alone, IL-6 (50 ng/mL), Dex (1 μmol/L), or IL-6 (50 ng/mL) + Dex (1 μmol/L) for 16 hours. Lysates were immunoprecipitated with anti-CDK2, anti-CDK4, or anti-CDK6 polyclonal antibodies followed by Western immunoblotting with either the same antibodies or anti-p21 MoAb. Expression of cyclin E, cyclin D2, and pRB were analyzed by immunoprecipitation and Western immunoblotting using the same cell lysates.
The binding of p21 to CDK2, CDK4, and CDK6 in these cell lysates was investigated using immunoprecipitation and Western blotting (Fig 5). p21 was constitutively bound to CDK2, CDK4, and CDK6. This binding was decreased by IL-6 and increased by Dex; in addition, IL-6 inhibited the upregulation in binding triggered by Dex. These changes parallelled changes in p21 protein expression in lysates from cells cultured under similar conditions (Fig 2).
Cyclin E and cyclin D2 were constitutively expressed in OCI-My5 MM cells, and their expression was not altered by IL-6 and/or Dex (Fig 5). Finally, both phosphorylated pRB (110 kD) and dephosphorylated pRB (105 kD) were observed in OCI-My5 MM cells cultured in media alone (Fig 5). IL-6 increased phosphorylation of pRB, whereas Dex triggered dephosphorylation of pRB. IL-6 inhibited Dex-induced dephosphorylation of pRB.
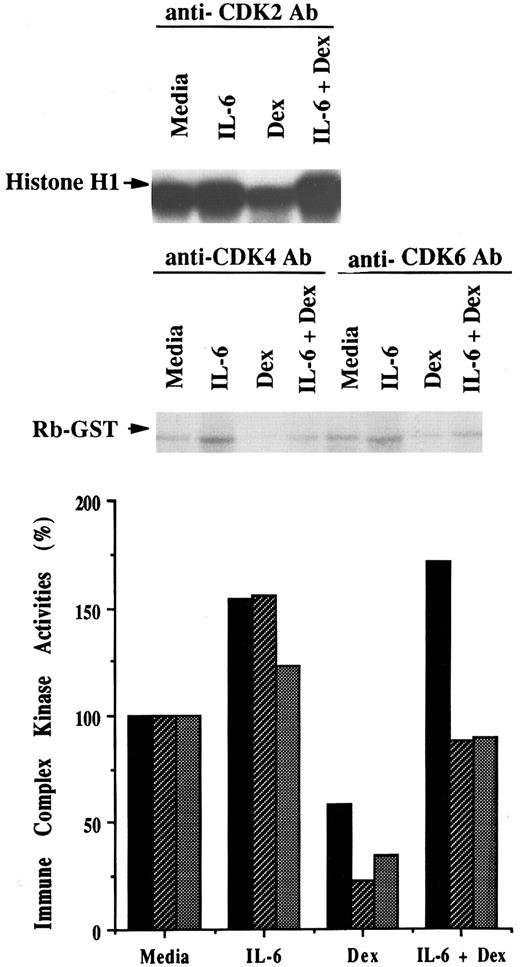
Effects of Dex and/or IL-6 on CDK2, CDK4, and CDK6 protein kinase activities.Given the effects of Dex and/or IL-6 on binding of p21 to CDK2, CDK4, and CDK6 described above, we next examined effects of Dex and/or IL-6 on CDK2-, CDK4-, and CDK6-associated kinase activities in OCI-My5 MM cells (Fig 6). Because p21 is known to induce dephosphorylation of pRB through suppression of CDK2-cyclin E complex as well as CDK4-cyclin D and CDK6-cyclin D complexes, cell lystates were immunoprecipitated with anti-CDK2, anti-CDK4, or anti-CDK6 antibodies; the CDK2- and CDK4 or CDK6-associated immune complexes were assayed for kinase activities by using histone H1 protein or purified GST-RB fusion protein substrates, respectively. Relative to cultures with media alone, IL-6 increased kinase activities of these complexes, as evidenced by phosphorylation of histone H1 or GST-RB fusion protein. In contrast, Dex inhibited activity of these kinases compared with media alone. IL-6 blocked the Dex-related downregulation of these kinase activities, corresponding to its affects on cell cycle distribution, p21 expression, and p21 binding to CDKs.
Effects of Dex and/or IL-6 on CDK2, CDK4, and CDK6 protein kinase activities. OCI-My5 MM cells were cultured with media alone, IL-6 (50 ng/mL), Dex (1 μmol/L), or Dex (1 μmol/L) and IL-6 (50 ng/mL). Total lysates were prepared 16 hours later and immunoprecipitated with anti-CDK2, anti-CDK4, or anti-CDK6 polyclonal Abs. Histone H1 or GST-RB fusion protein was used as substrate for kinase activity assays of CDK2 immune complex or of CDK4 and CDK6 immune complexes, respectively. The intensity was quantitated with densitometry and relative kinase activities of CDK2 (▪), CDK4 (▨), and CDK6 () compared in cells cultured in media, IL-6, Dex, or both IL-6 and Dex.
Effects of Dex and/or IL-6 on CDK2, CDK4, and CDK6 protein kinase activities. OCI-My5 MM cells were cultured with media alone, IL-6 (50 ng/mL), Dex (1 μmol/L), or Dex (1 μmol/L) and IL-6 (50 ng/mL). Total lysates were prepared 16 hours later and immunoprecipitated with anti-CDK2, anti-CDK4, or anti-CDK6 polyclonal Abs. Histone H1 or GST-RB fusion protein was used as substrate for kinase activity assays of CDK2 immune complex or of CDK4 and CDK6 immune complexes, respectively. The intensity was quantitated with densitometry and relative kinase activities of CDK2 (▪), CDK4 (▨), and CDK6 () compared in cells cultured in media, IL-6, Dex, or both IL-6 and Dex.
Effects of IFN-γ and/or IL-6 on cell cycle distribution and p21 protein expression in patient MM cells.Because IFN-γ suppresses the growth of epidermoid carcinoma cells by upregulating p21 transcription,31 we studied the effects of IFN-γ on cell cycle distribution (Table 4) and p21 protein expression (Fig 7) in the MM cells of patient no. 1. Cell cycle distribution did not significantly change during 48 hours of culture of the MM cells of patient no. 1 in media alone. As was noted above with Dex, IFN-γ increased the percentage of cells in G1 (peak 10% increments at 16 hours). In contrast, IL-6 downregulated the fraction of cells in G1 and upregulated those in S by 5% to 10%. Finally, as was observed with Dex + IL-6, the cell cycle distribution in the presence of both IFN-γ and IL-6 was similar to that noted in cultures with IL-6 alone.
Time-Dependent Effects of INF-γ and/or IL-6 on Cell Cycle Distribution of the Cells of Patient No. 1
| . | Time (h) . | |||||||
|---|---|---|---|---|---|---|---|---|
| . | 0 . | 2 . | 4 . | 8 . | 16 . | 24 . | 36 . | 48 . |
| Media | 80.1 | 79.5 | 79.6 | 80.2 | 81.5 | 82.9 | 83.4 | 83.8 |
| IL-6 (50 ng/mL) | 80.1 | 81.8 | 80.2 | 77.5 | 74.3 | 74.1 | 72 | 73.3 |
| INF-γ (10 ng/mL) | 80.1 | 81.7 | 86.7 | 89.5 | 92.6 | 92.5 | 92.2 | 92 |
| IL-6 (50 ng/mL) + INF-γ (10 ng/mL) | 80.1 | 82.6 | 81.9 | 80.3 | 78.5 | 77.6 | 75.3 | 75.2 |
| . | Time (h) . | |||||||
|---|---|---|---|---|---|---|---|---|
| . | 0 . | 2 . | 4 . | 8 . | 16 . | 24 . | 36 . | 48 . |
| Media | 80.1 | 79.5 | 79.6 | 80.2 | 81.5 | 82.9 | 83.4 | 83.8 |
| IL-6 (50 ng/mL) | 80.1 | 81.8 | 80.2 | 77.5 | 74.3 | 74.1 | 72 | 73.3 |
| INF-γ (10 ng/mL) | 80.1 | 81.7 | 86.7 | 89.5 | 92.6 | 92.5 | 92.2 | 92 |
| IL-6 (50 ng/mL) + INF-γ (10 ng/mL) | 80.1 | 82.6 | 81.9 | 80.3 | 78.5 | 77.6 | 75.3 | 75.2 |
Patient no. 1 MM cells (5 × 105) were cultured with 10% FBS RPMI-1640 media alone, IL-6, Dex, or IL-6 + Dex. Cells (1 × 106) were harvested at intervals, washed, stained with PI, and analyzed using FACS. Values are the percentage of cells in G1 phase.
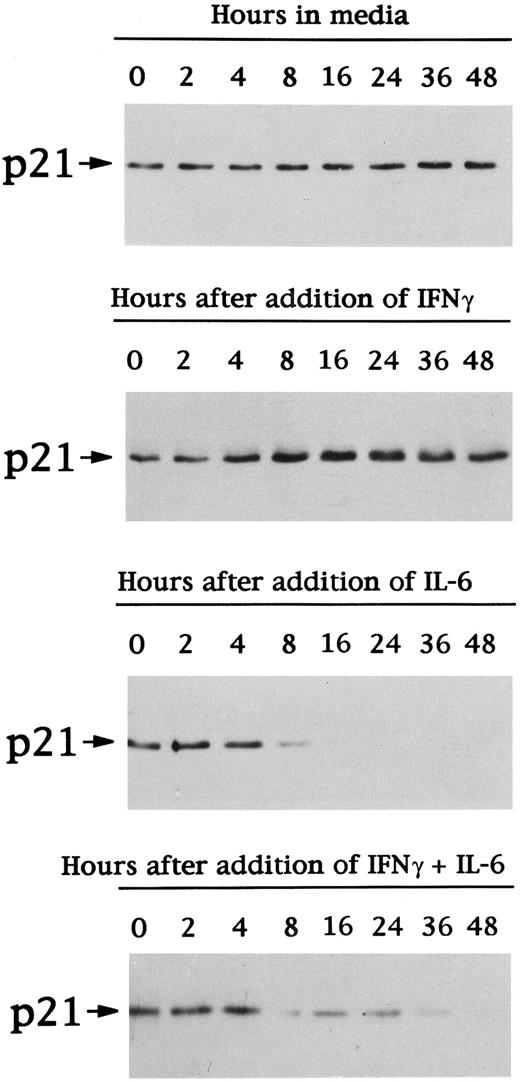
Effects of IFN-γ and/or IL-6 on expression of p21 protein in patient MM cells. Total lysates from MM cells of patient no. 1 were prepared before (time 0) and after 2, 4, 8, 16, 24, 36, and 48 hours of culture with 10% FBS-RPMI-1640 media, IFN-γ (10 ng/mL), IL-6 (50 ng/mL), or IFN-γ (10 ng/mL) + IL-6 (50 ng/mL). Total cell lysates were immunoprecipitated with anti-p21 polyclonal Ab, followed by Western immunoblotting with anti-p21 MoAb.
Effects of IFN-γ and/or IL-6 on expression of p21 protein in patient MM cells. Total lysates from MM cells of patient no. 1 were prepared before (time 0) and after 2, 4, 8, 16, 24, 36, and 48 hours of culture with 10% FBS-RPMI-1640 media, IFN-γ (10 ng/mL), IL-6 (50 ng/mL), or IFN-γ (10 ng/mL) + IL-6 (50 ng/mL). Total cell lysates were immunoprecipitated with anti-p21 polyclonal Ab, followed by Western immunoblotting with anti-p21 MoAb.
The kinetics of p21 protein expression in the MM cells of patient no. 1 was next examined under the same culture conditions (Fig 7). p21 protein expression in MM cells cultured in media alone was constant during the 48 hours of culture. IFN-γ increased and IL-6 decreased p21 protein expression; p21 protein expression in the presence of both IL-6 and IFN-γ was similar to that noted in cultures with IL-6 alone.
DISCUSSION
In the present study, we found p21 protein to be constitutively expressed in the majority of MM cells and to be undetectable in normal B cells and BMMCs. This may appear counterintuitive because p21 inhibits cellular proliferation in both p53-dependent and -independent mechanisms.32,33 However, p21 protein is also highly expressed in other malignancies, including glioblastoma, acute myelogenous leukemia (AML), and non–small-cell lung cancer.34-36 Morevoer, p21 gene is rarely mutated or deleted in cancers,37,38 and p21-deficient mice do not develop tumors at frequencies higher than their normal littermates.39 Interestingly, ectopic p21 expression has been shown to confer resistance to apoptosis40 and patients whose AML cells overexpress p21 protein demonstrate chemotherapy resistance.35 The constitutive p21 expression MM cells may therefore also result in resistance to apoptosis and related slow clinical progression of disease and chemoresistance.
Although p21 can be induced by p53, p21 protein expression in MM cells in this study was not related to the status of p53. This finding is consistent with reports in other malignancies that also show that overexpression of p21 is not related to the status of p5334-36 and suggests the existence of alternative mechanisms for induction of p21. Indeed, p21 is hypothesized to protect from p53-dependent apoptosis by induction of cell-cycle arrest and subsequent DNA repair.41,42 Moreover, other factors that induce apoptosis in a p53-independent mechanism, such as Dex at high doses,43 may also induce p21 and related cell cycle arrest at lower doses.23 Of interest, the expression of p27 protein in MM cells in our study was not related to p21 protein expression, suggesting that p21 and p27 may have different inducers or suppressors, even though they have high homology and similar function.
In the present report, we also demonstrated that both Dex and IFN-γ inhibited, whereas IL-6 facilitated, cell cycle progression from G1 to S phase. Dex has previously been reported to induce growth arrest and induce apoptosis in MM cells9; however, inhibition of cell proliferation by Dex occurs before induction of apoptosis,44 and the dose of Dex required to inhibit cell proliferation is at least 10-fold less than that required to induce programmed cell death.23 In our study, 0.1 to 1 μmol/L Dex suppressed growth, as evidenced by G1 growth arrest at 16 hours. IL-6 inhibited Dex-induced G1 growth arrest only in IL-6–responsive MM cells. Moreover, in this study, IFN-γ showed effects on MM cells similar to Dex, namely induction of G1 growth arrest.
The effects on cell cycle of Dex and IL-6 were correlated with changes in p21 expression. Specifically, in our study, Dex upregulated, whereas IL-6 downregulated, p21 expression in MM cells in a time- and dose-dependent fashion. Upregulation of p21 was temporally associated with G1 growth arrest induced by Dex, whereas downregulation of p21 triggered by IL-6 corresponded with G1 to S transition. These changes occurred only in IL-6–responsive MM cell lines and patient cells. It is of great interest that IL-6 inhibited the effects of Dex and IFN-γ on both cell cycle distribution and p21 expression in MM cells. Both IL-6 and Dex can regulate a variety of transcription factors, ie, via AP-1,24,45-47 and it is therefore possible that Dex and IL-6 may have opposing affects on gene transcription. Alternatively, Chin et al31 have demonstrated a sis-inducible element (SIE) in the p21 promotor that is a binding site for STAT proteins. In fact, IFN-γ has been shown to upregulate transcription of p21 gene, coupled with activation of STAT1, in epidermal cancer cells.31 STAT-3 is activated by IL-6,24 and it is therefore possible that distinct patterns of activation of STAT cascades by IL-6 and IFN-γ in MM cells may contribute to regulation of p21 transcription. Finally, IL-6 neither downregulated p21 nor blocked the upregulation of p21 induced by Dex in IL-6–nonresponsive MM cells, suggesting a block of upstream IL-6 signaling.
After showing that p21 was expressed in MM cell lines and MM patient cells, we next characterized p21 function, first in immunoprecipitation and immunoblotting assays with other known cell cycle regulatory proteins and then by kinase activity assays. In our study, p21 was constitutively bound to CDK2, CDK4, and CDK6 in MM cells; this binding was upregulated by Dex and decreased by IL-6, reflecting changes in p21 protein. Increased expression of p21 protein inhibits cyclin D-CDK4, cyclin D-CDK6, and cyclin E-CDK2 complexes and increases dephosphorylation of pRB, resulting in growth suppression.26 In this study, Dex-related increases in p21 in MM cells were associated with dephosphorylation of pRB and growth arrest; conversely, decreases in p21 were associated with phosphorylation of pRB and G1 to S phase transition. The changes in binding of p21 to CDK2, CDK4, and CDK6 triggered by Dex, as well as related dephosphorylation of pRB, were also blocked by IL-6.
To confirm the functional significance of Dex and/or IL-6–induced alterations in p21 on CDK2, CDK4, and CDK6, we measured kinase activities of p21-CDK2-cyclin E, p21-CDK4-cyclin D, and p21-CDK6-cyclin D immune complexes, which are known to affect phosphorylation of pRB. Activity of these kinases in the presence of IL-6 and/or Dex mirrored the results of immunoprecipitation and immunoblotting studies of p21 with CDK2, CDK4, and CDK6. Specifically, kinase activities of the immune complexes were upregulated by IL-6 and downregulated by Dex. Moreover, IL-6 overcame the downregulation of these kinases in the presence of Dex.
This study therefore demonstrates that changes in cell cycle distribution in MM cells triggered by Dex, IL-6, and IFN-γ correlate with changes in p21 protein expression and implicate p21 in coupling of Dex-, IL-6–, and IFN-γ–related signals to G1 cell cycle regulation in MM cells. Further studies of the function of p21 in tumor cells, as well as mechanisms regulating its expression, may not only enhance our understanding of mechanisms of cell cycle control in MM cells, but also suggest innovative therapies for MM that target p21.
Supported by National Institutes of Health Grant No. CA 50947.
Address reprint requests to Kenneth C. Anderson, MD, Dana-Farber Cancer Institute, 44 Binney St, Boston, MA 02215.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal