Abstract
The granulocyte-macrophage colony-stimulating factor receptor (GM-CSFR) is a potential target for toxin-directed therapy, because it is overexpressed on many leukemias and solid tumors and apparently not on stem cells. To investigate the potential therapeutic use of GM-CSF toxins, we fused human GM-CSF to truncated forms of either Pseudomonas exotoxin (PE) or diphtheria toxin (DT) and tested the cytotoxicity of the resulting GM-CSF–PE38KDEL and DT388–GM-CSF on human gastrointestinal (GI) carcinomas and leukemias. Toward gastric and colon cancer cell lines, GM-CSF–PE38KDEL was much more cytotoxic than DT388–GM-CSF, with IC50s (concentration resulting in 50% inhibition of protein synthesis) of 0.5 to 10 ng/mL compared with 4 to 400 ng/mL, respectively. In contrast, toward leukemia lines and fresh bone marrow cells DT388–GM-CSF was more cytotoxic than GM-CSF–PE38KDEL. The cytotoxicity of both GM-CSF–PE38KDEL and DT388–GM-CSF toward the human cells was specific, because it could be competed by an excess of GM-CSF. Binding studies indicated that human GM-CSF receptors were present on all of the human GI and leukemic cell lines tested, at levels of 540 to 3,700 sites per cell (kd = 0.2 to 2 nmol/L), and the number of sites per cell did not correlate with the cell type. A similar pattern of cytotoxicity was found with recombinant immunotoxins binding to the transferrin receptor, in that anti-TFR(Fv)–PE38KDEL was much more cytotoxic than DT388–anti-TFR(Fv) toward GI cells, but both were similar in their cytotoxic activity toward leukemia cells. The fact that PE is more effective than DT in killing GI but not leukemic tumor cells targeted by GM-CSF indicates a fundamental difference in the way PE or DT gains access to the cytosol in these cells. GM-CSF–PE38KDEL and DT388–GM-CSF deserve further evaluation as possible treatments for selected tumors.
GRANULOCYTE-MACROPHAGE colony-stimulating factor (GM-CSF) is a cytokine responsible for the growth, differentiation, and functional enhancement of granulocytes and macrophages.1-3 Human GM-CSF is 127 amino acids long and binds to hematopoietic cells via high-affinity (kd = 10 to 50 pmol/L) receptors composed of α and β subunits.4-8 GM-CSF is used to attenuate the myelosuppressive effects of chemotherapy in the treatment of not only hematologic malignancies but also solid tumors,9 and it has been shown that GM-CSF usually does not stimulate the growth of solid tumors.10 However, like leukemias,11 solid tumors, including renal, lung, breast, and gastrointestinal carcinomas, also express GM-CSF receptors (GM-CSFRs), at least the low affinity α component (kd = 0.7 to 2 nmol/L).12-16
Recent studies have shown that GM-CSFR is absent on the most immature hematopoietic progenitors but increases in expression during maturation.17 This finding suggests that the GM-CSFR may be a useful target for recombinant toxins or immunotoxins, which contain a cell binding protein linked to a protein toxin. Murine GM-CSF has recently been fused to truncated diphtheria toxin and the resulting DT390mGM-CSF was cytotoxic to murine GM-CSFR–bearing cells.18 A chemical conjugate of human GM-CSF with saporin was shown to kill mouse cells transfected with the human receptor.19 To determine the utility of GM-CSFR as a means to target human hematologic and solid tumors, we fused human GM-CSF to truncated forms of Pseudomonas exotoxin (PE) or diphtheria toxin (DT).
PE is a 66-kD protein that, like DT, kills cells by binding to a receptor, internalizing via a coated pit, translocating its active fragment into the cytosol, and enzymatically ADP-ribosylating elongation factor-2.20,21 The x-ray crystallographic structure of PE indicates three major domains, and mutational analysis has elucidated which domains are responsible for the several steps necessary to kill cells.22,23 Domain Ia, which is composed of amino acids 1 through 252, functions to bind the toxin to the PE receptor. Domain III (amino acids 400 through 613) contains the enzymatic activity that ADP-ribosylates EF2. Domain II (amino acids 253 through 364) undergoes proteolytic processing and is responsible for translocating to the cytosol the 37-kD carboxyl terminus of PE that contains the ADP ribosylating activity. DT also undergoes proteolytic processing,24 but its amino terminus contains the ADP-ribosylating activity and is translocated to the cytosol. Accordingly, in chimeric DT-containing toxins, the ligand replaces the toxin's binding domain at the carboxyl terminus. Conversely, in PE-containing chimeric toxins, the ligand replaces the toxin's binding domain at the amino terminus. The truncated form of PE used in the present study ends in KDEL, which has been shown to improve the cytotoxicity of PE-containing toxins and to increase binding of the toxin fragment to the KDEL receptor, which probably transports it to the endoplasmic reticulum, where it can translocate to the cytosol.25-27
MATERIALS AND METHODS
Plasmid construction.The polymerase chain reaction (PCR) was performed using a PCR kit from Perkin Elmer Cetus (Norwalk, CT). Denaturation temperature was 94°C for 1 minute, annealing was at 55°C for 2 minutes, and polymerization was at 72°C for 3 minutes, with 10 seconds of extension per cycle. Plasmids were sequenced using an Applied Biosystems Taq Dyedeoxy cycle-sequencing kit and an automated sequencer (Applied Biosystems, Foster City, CA). The cDNA encoding human GM-CSF and containing Nde I and HindIII restriction sites at the ends was obtained from a human spleen cDNA library (Clontech, Palo Alto, CA) using primers BK-134 (5′-gcc-tgc-agc-cat-atg-gca-ccc-gcc-cgc-tcg-ccc-agc-ccc-3′) and BK144 (3′-ctg-acg-acc-ctc-ggt-cag-gtc-ctc-att-cga-act-taa-gcc-5′). The 0.41-kb Nde I-HindIII fragment was then ligated into the 3.0-kb Nde I-HindIII fragment of pRKL4.28 The resulting plasmid, pRKGM2, contained the reported GM-CSF–encoding sequence,6 except for mutations of codons 83 (cac→cgc), 86 (cag→cgg), and 97 (gca→gcg). The mutations at codons 83 and 86 were repaired using revertant primers that separately amplified codons 1 through 87 and 79 through 127. Primers for the first amplification were BK-145 (5′-gga-gat-ata-cat-atg-gca-cca-gca-cga-tcg-cca-agc-cca-agc-acg-cagccc-t gg-3′) and BK-147 (3′-ggg-aac-tgg-tac-tac-cga-tcg-gtg-atg-ttc-gtc-gtg-5′), and for the second amplification were BK-146 (5′-atg-gct-agc-cac-tac-aag-cag-cac-tgc-cct-cca-acc-3′) and BK-148 (3′-ctg-acg-acc-ctc-ggt-cat-gtc-ctt-cga-aga-act-taa-5′). The two overlapping fragments were used as a template for amplification with BK-145 and BK-148. The 0.39-kb Nde I-HindIII fragment of the final amplification product was then ligated to the 4.5-kb fragment of pRKB3F,29 resulting in pRKGM9K, which had the correct sequence encoding GM-CSF–PE38KDEL. The 0.39-kb Nde I-HindIII fragment from pRKGM9K was then ligated to the 4.2-kb Nde I-HindIII fragment of pVCDT1-IL2,30 resulting in pRKDTGM, which encodes DT388–GM-CSF. To make HB9K, encoding anti-TFR(Fv)-PE38KDEL, the 0.35-kb Nde I-BamHI and 0.35-kb BamHI-HindIII fragments of plasmid pJBDT1-anti-TFR(Fv)31 were ligated to the 4.1-kb Nde I-HindIII fragment of pRK749K.32
Protein expression and purification.The method for expressing pRKGM9K, pRKDTGM, pRKHB9K, and pJBDT1-anti-TFR(Fv) and purifying the respective recombinant toxins GM-CSF–PE38KDEL, DT388–GM-CSF, anti-TFR(Fv)-PE38KDEL, and DT388-anti-TFR(Fv) differed slightly from the protocol previously reported.33Escherichia coli BL21/λDE3 cells34 were transformed with each plasmid and grown overnight on LB-ampicillin plates. The transformed cells were cultured in superbroth containing 5 g/L glucose, 1.4 mmol/L MgSO4, and 100 μg/mL ampicillin. At an OD650 of 2 to 3.5, protein synthesis was induced for 90 to 120 minutes with 1 mmol/L isopropyl-B-D-thiogalactopyranoside. The harvested cell paste was resuspended using a Tissuemizer tip (Thomas, Swedesboro, NJ) in TES buffer (50 mmol/L Tris, pH 8, 100 mmol/L NaCl, and 20 mmol/L EDTA) containing 180 μg/mL lysozyme. After incubating at 22°C for 1 hour, the cells were resuspended again and centrifuged at 27,000g for 50 minutes. The pellet was washed by resuspension and centrifugation three or four times with TES buffer containing 2.5% Triton-X-100 and then four times with TES. The inclusion bodies were resuspended in 5 to 10 mL of denaturation buffer (7 mol/L guanidine:HCl, 0.1 mol/L Tris, pH 8.0, and 5 mmol/L EDTA) by sonication or tissuemizing and diluted to a protein concentration of 10 mg/mL. The protein was reduced with dithioerythritol (65 mmol/L) for 4 to 24 hours at 22°C and rapidly diluted in a thin stream into refolding buffer (0.1 mol/L Tris, pH 8.0, 0.5 mol/L arginine:HCl, 2 mmol/L EDTA, and 0.9 mmol/L oxidized glutathione). After incubating at 10°C for 36 to 72 hours, the refolding buffer was either diluted 10-fold with water or dialyzed against 0.02 mol/L Tris, pH 7.4, 1 mmol/L EDTA, and 0.1 mol/L urea. The filtered protein was then purified by Qsepharose and MonoQ (Pharmacia, Piscataway, NJ) anion exchange and finally by sizing chromatography. The yield of purified active monomeric protein was 7.5% to 10% of total denatured recombinant protein.
Cytotoxicity assay.N87 gastric carcinoma cells were obtained from Dr R. King (Georgetown University, Washington, DC),35 HUT-102 adult T-cell leukemia (ATL) cells were obtained from Dr T. Waldmann (National Institutes of Health), and the other cell lines were available from ATCC (Rockville, MD). A total of 1.5 × 104 cells/well were plated in 96-well plates; 24 hours later, toxin or control molecules were added and incubated for 48 hours in final volumes of 200 μL. The cells were pulsed for 4 to 6 hours with [3H]-leucine 1 μCi/well, harvested, and counted. Bone marrow mononuclear cells were obtained from a patient with lymphocytic leukemia and from a normal donor for allogeneic bone marrow transplantation by Ficoll centrifugation, as described.36 The marrow mononuclear cells (0.4 to 1 × 106/well) were incubated for 60 hours with toxin or control molecules in 100-μL aliquots of media consisting of 88% leucine-free RPMI, 2% RPMI, and 10% fetal bovine serum. The cells were then pulsed for 6 to 8 hours with [3H]-leucine at 2 μCi/well, harvested, and counted. The IC50 was the concentration of toxin required for 50% protein synthesis inhibition.
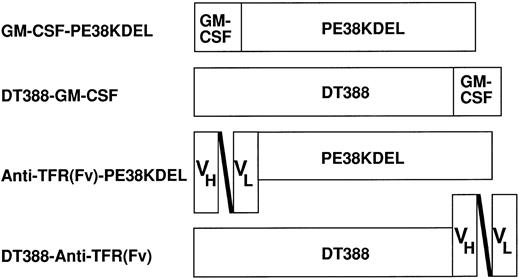
Schematic diagram of the recombinant toxins used. GM-CSF–PE38KDEL, encoded by pRKGM9K, contains the 127 amino acids of human GM-CSF followed by amino acids 253 through 364 and 381 through 608 of PE and then the sequence KDEL. DT388–GM-CSF, encoded by pRKDTGM, contains the first 388 amino acids of DT followed by human GM-CSF. Anti-TFR(Fv)–PE38KDEL and DT388–anti-TFR(Fv), encoded by pRKHB9K and pJBDT1–anti-TFR(Fv), respectively, contain the same toxin domains as the respective GM-CSF toxins, but the ligand is the single-chain Fv of an antitransferrin receptor antibody.
Schematic diagram of the recombinant toxins used. GM-CSF–PE38KDEL, encoded by pRKGM9K, contains the 127 amino acids of human GM-CSF followed by amino acids 253 through 364 and 381 through 608 of PE and then the sequence KDEL. DT388–GM-CSF, encoded by pRKDTGM, contains the first 388 amino acids of DT followed by human GM-CSF. Anti-TFR(Fv)–PE38KDEL and DT388–anti-TFR(Fv), encoded by pRKHB9K and pJBDT1–anti-TFR(Fv), respectively, contain the same toxin domains as the respective GM-CSF toxins, but the ligand is the single-chain Fv of an antitransferrin receptor antibody.
Binding assay.Clinical grade GM-CSF was purchased from Immunex (Seattle, WA) and desalted on a PD-10 column (Pharmacia, Piscataway, NJ), equilibrated, and eluted with phosphate-buffered saline (PBS). Na 125I (1 mCi; Amersham, Arlington Heights, IL) was added to a 100 μL volume of PBS containing GM-CSF (50 μg), sodium phosphate, pH 7.5 (150 mmol/L), and chloramine T (3.3 μg). After incubating for 2 minutes at 22°C, sodium metabisulfite (83 μg) was added and the [125I]–GM-CSF purified on a PD-10 (Pharmacia) column equilibrated and eluted with 0.2% human serum albumin in PBS. HB21 (also termed anti-TFR–IgG), the monoclonal antibody to the transferrin receptor,37 was labeled the same way except using 75 μg of anti-TFR–IgG and 10 μg of chloramine T. To determine the number of GM-CSFR sites per cell, cells were incubated in 96-well U-bottom plates in binding buffer (RPMI containing 0.1% bovine serum albumin and 0.2% NaN3) containing increasing concentrations of [125I]–GM-CSF with or without a 100-fold excess of unlabeled GM-CSF. After 30 minutes at 37°C, the cells were centrifuged and washed once with binding buffer and then counted. This method was easier and more reproducible than centrifuging the cells through n-butyl phthalate and counting the cell pellets. To determine the binding affinity of unlabeled toxins relative to that of GM-CSF, U937 cells (4.8 × 106/well) were plated in U-bottom 96-well plates in binding buffer and incubated with 1.2 nmol/L [125I]–GM-CSF with and without different concentrations of recombinant toxins. The relative binding affinity of the immunotoxins containing anti-TFR(Fv) was determined similarly using HUT-102 cells (8 × 105/well) and 0.1 nmol/L [125I] — anti-TFR–IgG.
RESULTS
The fact that GM-CSFR is overexpressed on solid tumors and leukemia cells but is undetectable on the earliest hematopoietic progenitor cells11-17 makes the GM-CSFR a potential target molecule. To determine whether GM-CSF can direct bacterial toxins to kill GM-CSF–expressing tumor cells, we fused GM-CSF to truncated forms of PE or DT and tested the resulting fusion toxins for cytotoxicity and binding.
Cytotoxicity of Recombinant Toxins Containing Human GM-CSF
| Cell Line . | Cell Type . | IC50 (ng/mL) ± SD . | |
|---|---|---|---|
| . | . | GM-CSF–PE38KDEL . | DT388–GM-CSF . |
| LS174T | Colon | 2.2 ± 0.7 | 70 ± 18 |
| SW403 | Colon | 0.9 ± 0.5 | 15 ± 0.6 |
| N87 | Gastric | 0.45 ± 0.2 | 3.7 ± 0.1 |
| HTB-103 | Gastric | 9.5 ± 7.5 | 400 ± 300 |
| HL60 | Promyelocytic | > 100 | 0.4 ± 0.2 |
| TF-1 | Erythroleukemia | 22 ± 8 | 0.02 ± 0.01 |
| U937 | Monocytic | 9.5 ± 5.5 | 0.04 ± 0.02 |
| Cell Line . | Cell Type . | IC50 (ng/mL) ± SD . | |
|---|---|---|---|
| . | . | GM-CSF–PE38KDEL . | DT388–GM-CSF . |
| LS174T | Colon | 2.2 ± 0.7 | 70 ± 18 |
| SW403 | Colon | 0.9 ± 0.5 | 15 ± 0.6 |
| N87 | Gastric | 0.45 ± 0.2 | 3.7 ± 0.1 |
| HTB-103 | Gastric | 9.5 ± 7.5 | 400 ± 300 |
| HL60 | Promyelocytic | > 100 | 0.4 ± 0.2 |
| TF-1 | Erythroleukemia | 22 ± 8 | 0.02 ± 0.01 |
| U937 | Monocytic | 9.5 ± 5.5 | 0.04 ± 0.02 |
Cells were plated in 96-well plates at 1.5 × 104/well and incubated at 37°C for 24 hours. The cells were then incubated with recombinant toxins for 24 to 48 hours and [3H]-leucine for 4 to 6 hours.
Cytotoxicity and specificity of GM-CSF toxins on target cells. The colon lines SW403 (A) and LS174T (B) were incubated with GM-CSF–PE38KDEL and DT388–GM-CSF was incubated with the leukemia lines HL60 (C) and U937 (D) in the presence (•, ▴) or absence (○, ▵) of 5 μg/mL of GM-CSF. The cells were pulsed with [3H]-leucine and harvested, and the leucine incorporation was determined.
Cytotoxicity and specificity of GM-CSF toxins on target cells. The colon lines SW403 (A) and LS174T (B) were incubated with GM-CSF–PE38KDEL and DT388–GM-CSF was incubated with the leukemia lines HL60 (C) and U937 (D) in the presence (•, ▴) or absence (○, ▵) of 5 μg/mL of GM-CSF. The cells were pulsed with [3H]-leucine and harvested, and the leucine incorporation was determined.
Preparation of recombinant GM-CSF toxins.Figure 1 shows schematic diagrams of GM-CSF–PE38KDEL and DT388–GM-CSF. As confirmed by the DNA sequence analysis, the first molecule contains the 127 amino acid human GM-CSF ligand at the amino terminus of the truncated toxin, which consists of amino acids 253 through 364 and 381 through 608 of PE, followed by the KDEL carboxyl terminus. DT388–GM-CSF contains the first 388 amino acids of DT followed by human GM-CSF. GM-CSF was placed at the amino terminus of truncated PE and at the carboxyl terminus of truncated DT to replace the binding domains that are normally present in those positions and because a free carboxyl terminus of PE and a free amino terminus of DT is necessary for cytotoxicity.30 38 To properly fold the recombinant toxins, each of which contains 6 cysteine residues, the insoluble inclusion body protein was denatured, reduced, and refolded in a redox buffer as described in Materials and Methods. Each protein could be purified to near homogeneity by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (gel not shown) in a yield of 10% of the total recombinant protein renatured. The purified recombinant toxins were then tested for cytotoxicity and binding.
Cytotoxicity of GM-CSF toxins.To determine the sensitivity of leukemia and GI carcinomas to GM-CSF toxins, the cells were incubated with recombinant toxins and [3H]-leucine incorporation was measured. The IC50s, the concentrations of toxin necessary for 50% inhibition of protein synthesis, are listed in Table 1. For GM-CSF–PE38KDEL, the IC50s on GI carcinomas ranged from 0.45 ng/mL on N87 to 9.5 ng/mL on HTB-103 cells. Leukemia cells were less sensitive to GM-CSF–PE38KDEL, with IC50s ranging from 9.5 ng/mL on U937 monocytic leukemia cells to greater than 100 ng/mL on HL60 promyelocytic leukemia cells. In contrast, DT388–GM-CSF was much more cytotoxic to leukemia cells than to GI carcinomas, with IC50s ranging from 0.02 ng/mL to 0.4 ng/mL on the leukemia lines compared with 3.7 ng/mL to 400 ng/mL on the GI carcinoma lines. Thus, GM-CSF–PE38KDEL was much more cytotoxic than DT388–GM-CSF on the GI carcinoma lines, whereas the reverse was true on the leukemia lines.
Cytotoxic specificity of the recombinant toxins containing GM-CSF.To determine whether the cytotoxicity of the recombinant GM-CSF toxins was specific in requiring internalization through the GM-CSF receptor, cytotoxicity experiments were performed where the binding of the chimeric toxins was competed by an excess of GM-CSF (2.5 to 5 μg/mL). Figure 2 shows representative cytotoxicity curves for GM-CSF–PE38KDEL and DT388–GM-CSF in the presence and absence of an excess of GM-CSF. It was found that an excess of GM-CSF prevented the cytotoxic activity of both recombinant toxins, indicating that their cytotoxic activity required binding to the GM-CSF receptor. Such specificity was also shown by GM-CSF–PE38KDEL for N87, HTB-103, TF-1, and U937 cells and by DT388–GM-CSF for TF-1, LS174T, and SW403 cells (data not shown). To determine whether the cytotoxicity of the GM-CSF toxins was only due to their binding to the GM-CSFR and did not require the action of the toxin domains, the cells were incubated with and without 5 μg/mL of GM-CSF. In Fig 2, the points on the y-axis of each curve, where the toxin concentration equals 0, show the leucine incorporation in cells with and without 5 μg/mL of GM-CSF. In none of the seven cell lines did GM-CSF alone result in greater than 50% inhibition of protein synthesis. Thus, the cytotoxicity of GM-CSF–PE38KDEL and DT388–GM-CSF required both binding to the GM-CSFR and also internalization and action of the toxin domains. Interestingly, the leukemic cell lines HL60 (Fig 2C) and TF-1 (data not shown) showed significant (but <twofold) stimulation with 5 μg/mL of GM-CSF in the absence of toxin, but the solid tumors showed no significant stimulatory response to GM-CSF.
Quantitation of GM-CSFR on the target cells.The number of GM-CSFR sites per cell were quantitated by radiolabeled binding assay using [125I]–GM-CSF on the cell lines to determine whether cytotoxic activity correlated with receptor expression. Representative Scatchard plots are shown in Fig 3. The binding assay was designed to determine the number of low-affinity sites, which outnumber the small number of high-affinity sites.4-8 As shown in Table 2, the number of GM-CSFR sites per cell varied somewhat from assay to assay, but the average values varied from 500 sites per cell for HTB-103 gastric cells to 3,700 sites per cell for TF-1 cells. The kds for the seven cell lines ranged from 0.2 to 1.9 nmol/L, consistent with low-affinity binding sites. Together with the cytotoxicity data from Table 1, it can be seen that cytotoxicity did correlate with the number of GM-CSFR sites per cell when examining one type of cell at a time. For example, for either toxin toward the GI carcinoma lines, SW403 and N87 were more sensitive than LS174T and HTB-103 was least sensitive, matching the order of their GM-CSFR expression. Also, of the three leukemia lines, HL60 had the least numbers of sites per cell and was less sensitive to either toxin than were TF-1 or U937 cells. However, GM-CSF–PE38KDEL was more cytotoxic toward the GI lines than the leukemia lines, and DT388–GM-CSF was more cytotoxic toward the leukemia lines than the GI lines. Thus, for both recombinant toxins the difference in their cytotoxic activity toward leukemia and solid tumor cells was likely due to fundamental differences between these cell types unrelated to the numbers of receptors expressed.
Scatchard plots of [125I]–GM-CSF binding to target cells. In (A), N87 cells (1.3 × 106/200 μL/well) were incubated with 0.15, 0.3, 0.6, 1.2, 2.4, and 4.8 nmol/L [125I]–GM-CSF (5.7 μCi/μg) in RPMI containing 0.1% bovine serum albumin and 0.2% NaN3 for 30 minutes, washed, and counted. HL60 cells (B) were assayed similarly at 2.2 × 106/well.
Scatchard plots of [125I]–GM-CSF binding to target cells. In (A), N87 cells (1.3 × 106/200 μL/well) were incubated with 0.15, 0.3, 0.6, 1.2, 2.4, and 4.8 nmol/L [125I]–GM-CSF (5.7 μCi/μg) in RPMI containing 0.1% bovine serum albumin and 0.2% NaN3 for 30 minutes, washed, and counted. HL60 cells (B) were assayed similarly at 2.2 × 106/well.
Expression of GM-CSFR on Human Cells
| Cell Line . | Sites/Cell . | kd (nmol/L) . |
|---|---|---|
| LS174T | 900 ± 700 | 1.7 ± 1 |
| SW403 | 3,050 ± 900 | 1.6 ± 0.5 |
| N87 | 2,400 ± 1,200 | 0.9 ± 0.7 |
| HTB-103 | 500 ± 200 | 0.4 ± 0.4 |
| HL60 | 540 ± 50 | 0.2 ± 0.01 |
| TF-1 | 3,700 ± 1,500 | 0.5 ± 0.05 |
| U937 | 3,500 ± 350 | 1.9 ± 0.2 |
| Cell Line . | Sites/Cell . | kd (nmol/L) . |
|---|---|---|
| LS174T | 900 ± 700 | 1.7 ± 1 |
| SW403 | 3,050 ± 900 | 1.6 ± 0.5 |
| N87 | 2,400 ± 1,200 | 0.9 ± 0.7 |
| HTB-103 | 500 ± 200 | 0.4 ± 0.4 |
| HL60 | 540 ± 50 | 0.2 ± 0.01 |
| TF-1 | 3,700 ± 1,500 | 0.5 ± 0.05 |
| U937 | 3,500 ± 350 | 1.9 ± 0.2 |
Cells were incubated at 37°C for 30 minutes with [125I]–GM-CSF with or without a 100-fold excess of GM-CSF in RPMI media containing 1 mg/mL bovine serum albumin and 0.2% NaN3. Unbound GM-CSF was removed from the cells either by centrifuging through n-butyl phthalate or by centrifuging and washing the cells with binding buffer.
Sensitivity of fresh bone marrow cells to GM-CSF toxins.To determine if the data on cell lines from patients with leukemia and GI cancer would apply to fresh human hematopoietic cells, bone marrow mononuclear cells from two donors were partially purified by Ficoll centrifugation and incubated with the recombinant toxins. The first sample was taken from normal marrow and the second from a patient whose marrow contained normal hematopoietic progenitor cells and was 50% involved with a B-cell small-cell lymphoma. As shown in Table 3, the marrow mononuclear cells were much more sensitive to DT388–GM-CSF compared with GM-CSF–PE38KDEL. In the normal marrow sample, the IC50 for DT388–GM-CSF was 2.1 ng/mL, compared with greater than 1,000 ng/mL for GM-CSF – PE38KDEL. The results were similar in the marrow contaminated with malignant B cells, with an IC50 of 0.9 ng/mL for DT388–GM-CSF and greater than 100 ng/mL for GM-CSF–PE38KDEL. Although these marrow samples contained differentiated cells, the cytotoxicity of DT388–GM-CSF appeared directed toward the hematopoietic progenitor cells, because fresh normal or malignant lymphocytes isolated from the peripheral blood were resistant (Table 3). To determine whether the cytotoxic activity of GM-CSF toxin towards the marrow cells was specific in requiring binding to the GM-CSFR, we simultaneously tested recombinant toxin containing PE38KDEL or DT388 but not GM-CSF. As shown in Fig 4A, PE38KDEL alone39 or DT388-IL230 showed no cytotoxic activity toward the normal marrow cells, and PE38KDEL was significantly less cytotoxic than GM-CSF–PE38KDEL. Thus, the cytotoxic activity of the recombinant toxins containing GM-CSF was not due to nonspecific internalization into the fresh marrow cells. As shown by the Y-axis of Fig 4B, in the absence of toxin, 20 ng/mL of GM-CSF resulted in a 25% increase in protein synthesis, indicating that the cytotoxic activity of the GM-CSF toxin was not only due to their binding to the GM-CSFR on the cells but also required action of the bacterial toxins after internalization. Finally, Fig 4B shows that the cytotoxic activity of DT388–GM-CSF could be competed by 20 ng/mL of GM-CSF, confirming that its cytotoxic activity required binding to the GM-CSFR on the fresh human hematopoietic progenitor cells.
Sensitivity of Fresh Hematopoietic Cells to Recombinant GM-CSF Toxins
| Sample . | IC50 (ng/mL) . | |
|---|---|---|
| . | GM-CSF–PE38KDEL . | DT388–GM-CSF . |
| Normal bone marrow | >1,000 | 2.1 |
| Marrow in lymphoma patient | >100 | 0.9 |
| Normal peripheral lymphocytes | >1,000 | >1,000 |
| Peripheral blood B leukemia | >1,000 | >1,000 |
| Sample . | IC50 (ng/mL) . | |
|---|---|---|
| . | GM-CSF–PE38KDEL . | DT388–GM-CSF . |
| Normal bone marrow | >1,000 | 2.1 |
| Marrow in lymphoma patient | >100 | 0.9 |
| Normal peripheral lymphocytes | >1,000 | >1,000 |
| Peripheral blood B leukemia | >1,000 | >1,000 |
Samples consisted of mononuclear cells obtained by Ficoll centrifugation and were incubated with recombinant toxins for 60 hours followed by [3H]-leucine for 4 to 6 hours.
Cyotoxicity of recombinant toxins towards normal bone marrow cells. Normal marrow mononuclear cells (106/0.1 mL well) were incubated with DT388–GM-CSF (▵) or GM-CSF–PE38KDEL (○) for 60 hours and pulsed with [3H]-leucine. In (A), the cells were incubated in parallel with the negative control molecules PE38KDEL (•) or DT388-IL2 (▴). In (B), the cells were also incubated with DT388–GM-CSF (▪) or GM-CSF–PE38KDEL (□) in the presence of 20 ng/mL of GM-CSF.
Cyotoxicity of recombinant toxins towards normal bone marrow cells. Normal marrow mononuclear cells (106/0.1 mL well) were incubated with DT388–GM-CSF (▵) or GM-CSF–PE38KDEL (○) for 60 hours and pulsed with [3H]-leucine. In (A), the cells were incubated in parallel with the negative control molecules PE38KDEL (•) or DT388-IL2 (▴). In (B), the cells were also incubated with DT388–GM-CSF (▪) or GM-CSF–PE38KDEL (□) in the presence of 20 ng/mL of GM-CSF.
Sensitivity of GI and leukemia cells to toxins carrying another ligand.To determine whether the difference in sensitivity of GI and leukemia cells to recombinant toxins was due to differences in the way the cells handle the GM-CSFR or due to differences between these cells that are unrelated to the GM-CSFR, they were incubated with recombinant toxins containing a different ligand. The ligand chosen was anti-TFR(Fv), which binds to the human transferrin receptor.31 As shown in Fig 1, anti-TFR(Fv)–PE38KDEL contains the exact same toxin domains as GM-CSF – PE38KDEL, and DT388–anti-TFR(Fv) contains the exact same toxin domains as DT388–GM-CSF. Table 4 lists the IC50s of these two recombinant immunotoxins toward the GI and leukemia cell lines. It can be seen that anti-TFR(Fv)–PE38KDEL was usually much more cytotoxic to the GI cell lines than the leukemia cell lines, with differences being as great as 200-fold. In contrast, DT388–anti-TFR(Fv) was usually more cytotoxic to the leukemia cells than to the GI carcinoma cells, with the differences being less than 10-fold. Thus, regardless of the targeting ligand, GI carcinomas were more sensitive than leukemias to PE38KDEL, and leukemias were more sensitive than GI carcinomas to DT388.
Sensitivity of Colon and Leukemia Lines to Recombinant Immunotoxins Binding to the Human Transferrin Receptor
| Cell Line . | Cell Type . | IC50 (ng/mL) ± SD . | |
|---|---|---|---|
| . | . | Anti-TFR(Fv)–PE38KDEL . | DT388–Anti-TFR(Fv) . |
| LS174T | Colon | 0.008 ± 0.003 | 0.4 ± 0.05 |
| SW403 | Colon | 0.02 ± 0.007 | 0.7 ± 0.3 |
| N87 | Gastric | 0.003 ± 0.0004 | 0.2 ± 0.07 |
| HTB-103 | Gastric | 0.002 ± 0.0006 | 0.2 ± 0.06 |
| HL60 | Promyelocytic | 0.4 ± 0.15 | 0.2 ± 0.015 |
| TF-1 | Erythroleukemia | 0.018 ± 0.006 | 0.13 ± 0.1 |
| U937 | Monocytic | 0.1 ± 0.09 | 0.13 ± 0.08 |
| Cell Line . | Cell Type . | IC50 (ng/mL) ± SD . | |
|---|---|---|---|
| . | . | Anti-TFR(Fv)–PE38KDEL . | DT388–Anti-TFR(Fv) . |
| LS174T | Colon | 0.008 ± 0.003 | 0.4 ± 0.05 |
| SW403 | Colon | 0.02 ± 0.007 | 0.7 ± 0.3 |
| N87 | Gastric | 0.003 ± 0.0004 | 0.2 ± 0.07 |
| HTB-103 | Gastric | 0.002 ± 0.0006 | 0.2 ± 0.06 |
| HL60 | Promyelocytic | 0.4 ± 0.15 | 0.2 ± 0.015 |
| TF-1 | Erythroleukemia | 0.018 ± 0.006 | 0.13 ± 0.1 |
| U937 | Monocytic | 0.1 ± 0.09 | 0.13 ± 0.08 |
Displacement analysis of recombinant toxins. In (A), U937 cells (4.5 × 106/well) were incubated with [125I]–GM-CSF and the indicated concentrations of GM-CSF (▪), GM-CSF–PE38KDEL (○), or DT388–GM-CSF (▵). In (B), HUT-102 cells (8 × 105/well) were incubated with [125I] — anti-TFR–IgG and the indicated concentrations of anti-TFR–IgG (▪), anti-TFR(Fv)–PE38KDEL (○), or DT388–anti-TFR(Fv) (▴). The cells were harvested and counted as in Fig 3.
Displacement analysis of recombinant toxins. In (A), U937 cells (4.5 × 106/well) were incubated with [125I]–GM-CSF and the indicated concentrations of GM-CSF (▪), GM-CSF–PE38KDEL (○), or DT388–GM-CSF (▵). In (B), HUT-102 cells (8 × 105/well) were incubated with [125I] — anti-TFR–IgG and the indicated concentrations of anti-TFR–IgG (▪), anti-TFR(Fv)–PE38KDEL (○), or DT388–anti-TFR(Fv) (▴). The cells were harvested and counted as in Fig 3.
Assessment of the relative affinities of the recombinant toxins.To compare the cytotoxic activities of GM-CSF–PE38KDEL or anti-TFR(Fv)–PE38KDEL with those of DT388–GM-CSF and DT388–anti-TFR(Fv), respectively, one must take into consideration quantitative differences in the binding of the ligands, depending on whether the ligand is located at the amino or carboxyl terminus of the toxin. To determine the relative binding affinity of the recombinant toxins, they were tested for their ability to displace radiolabeled ligand. In Fig 5A, the amount of [125I]–GM-CSF bound to U937 cells is shown as a function of increasing concentrations of either GM-CSF, GM-CSF–PE38KDEL, or DT388–GM-CSF. The EC50, the concentration necessary for 50% competition of [125I]–GM-CSF binding, was 3.9 nmol/L for GM-CSF, 41 nmol/L for GM-CSF–PE38KDEL, and 50 nmol/L for DT388–GM-CSF. In Fig 5B, the amount of [125I] — anti-TFR–IgG binding to HUT-102 cells is shown as a function of increasing concentrations of either anti-TFR–IgG, anti-TFR(Fv)–PE38KDEL, or DT388–anti-TFR(Fv). The EC50 was 13 nmol/L for anti-TFR–IgG, 12 nmol/L for anti-TFR(Fv)–PE38KDEL, and 29 nmol/L for DT388–anti-TFR(Fv). Table 5 was then constructed to display the IC50s for the recombinant toxins corrected for binding affinity. Lower values of IC50/EC50 would indicate more efficient killing of cells due to processes occurring after binding. Table 5 indicates that the IC50/EC50 ratios of GM-CSF–PE38KDEL are much less than those of DT388–GM-CSF toward GI carcinomas, whereas the reverse was true for leukemias. Similarly, the IC50/EC50 ratios of anti-TFR(Fv)–PE38KDEL are much less than those of DT388–anti-TFR(Fv) toward GI carcinomas, but not toward leukemias. Thus, regardless of the targeting ligand, PE38KDEL was more cytotoxic toward solid tumors and DT388 was more cytotoxic toward leukemias due to processes in those cells that occur after receptor binding.
Cytotoxicity of Recombinant Toxins Corrected for Binding Affinity
| Cell Line . | IC50 (pmol/L)/EC50 (nmol/L) . | |||
|---|---|---|---|---|
| . | GM-CSF–PE38KDEL . | DT388–GM-CSF . | Anti-TFR(Fv)–PE38KDEL . | DT388 – . |
| . | . | . | . | Anti-TFR(Fv) . |
| LS174T | 1 | 25 | 0.003 | 0.2 |
| SW403 | 0.4 | 5 | 0.007 | 0.35 |
| N87 | 0.2 | 1.3 | 0.001 | 0.1 |
| HTB-103 | 4 | 140 | 0.0007 | 0.1 |
| HL60 | >45 | 0.14 | 0.14 | 0.1 |
| TF-1 | 10 | 0.007 | 0.006 | 0.07 |
| U937 | 4 | 0.014 | 0.035 | 0.07 |
| Cell Line . | IC50 (pmol/L)/EC50 (nmol/L) . | |||
|---|---|---|---|---|
| . | GM-CSF–PE38KDEL . | DT388–GM-CSF . | Anti-TFR(Fv)–PE38KDEL . | DT388 – . |
| . | . | . | . | Anti-TFR(Fv) . |
| LS174T | 1 | 25 | 0.003 | 0.2 |
| SW403 | 0.4 | 5 | 0.007 | 0.35 |
| N87 | 0.2 | 1.3 | 0.001 | 0.1 |
| HTB-103 | 4 | 140 | 0.0007 | 0.1 |
| HL60 | >45 | 0.14 | 0.14 | 0.1 |
| TF-1 | 10 | 0.007 | 0.006 | 0.07 |
| U937 | 4 | 0.014 | 0.035 | 0.07 |
DISCUSSION
To determine the sensitivity of solid tumors and leukemias to recombinant toxins containing GM-CSF, we tested the sensitivity of such cell lines to GM-CSF–PE38KDEL and DT388–GM-CSF. We found that, although both recombinant toxins were specifically cytotoxic to both types of cells, solid tumor cells were more sensitive to GM-CSF–PE38KDEL and leukemia cells were more sensitive to DT388–GM-CSF. This was not a phenomenon peculiar to cell lines, because fresh human marrow progenitor cells behaved similar to the leukemia lines. Based on a similar pattern of cytotoxicity using the same toxins targeted to the transferrin receptor, it appears that these differences are due to inherent differences between solid tumors and leukemias in the way by which they handle the toxins intracellularly.
It is believed that cytotoxicity by PE requires proteolytic processing by Furin between amino acids 279 and 280 and that the carboxy terminal fragment is transported by the KDEL receptor from at least the transreticular Golgi toward the endoplasmic reticulum, from which it translocates to the cytosol.25,38,40,41 DT is also cleaved by Furin between arginine 193 and serine 194, and arginine 193 at the carboxyl terminus of the DT fragment A is necessary for translocation.24,40 However, this fragment is thought to translocate from the endosome to the cytosol via membrane insertion and passage through ion-conductive channels.42-44 Because both PE and DT are processed by the same enzyme, both toxins ultimately ADP ribosylate EF2 in the cytosol, and in both PE38KDEL and DT388 ADP ribosylation activity is not altered by the ligand to which the toxin is fused (data not shown). Thus, the difference in activity between GM-CSF–PE38KDEL and DT388–GM-CSF would not be due to differences in ADP-ribosylation activity. We therefore speculate from our data that leukemic cells differ from GI carcinoma cells in the intracellular transport or translocation of protein molecules. For example, in leukemic cells, a protein molecule may have a higher chance of entering the cytosol if it can translocate directly from the endosome than if it must be transported intracellularly. Conversely, in solid tumors, proteins may not easily translocate from endosomes and may require transport to an organelle such as the endoplasmic reticulum that has preexisting pores. It is also possible that leukemic and solid tumor cells differ in the function and location of lysosomes, which may degrade toxin molecules before they reach the cytosol. So far, our data with cell lines are consistent with that on fresh cells from patients with GM-CSFR+ leukemia (Rozemuller et al, manuscript submitted). Because one molecule of DT45 or PE (Willingham and Pastan, unpublished data) in the cytosol appears sufficient to kill a cell, it is very important to identify and attempt to overcome these potential impediments to translocation.
We have shown that human GM-CSF can be used to target truncated forms of either PE or DT to inhibit protein synthesis in receptor-bearing solid or hematopoietic tumor cells. Further development of these agents for the treatment of human tumors should include monkey toxicology studies, because murine GM-CSFR does not bind human GM-CSF.46 Such studies could determine whether cytotoxicity toward GM-CSFR+ normal cells spares stem cells and, if so, whether hematopoietic toxicity is low enough to allow the delivery of high doses. If so, such agents could prove useful clinically in the purging of bone marrow or stem cell autografts before transplantation or in the direct systemic treatment of human leukemia and solid tumors.
ACKNOWLEDGMENT
The authors acknowledge the technical assistance of R. Beers, I. Margulies, and V. Fogg, which contributed to aspects of this work.
Address reprint requests to Ira Pastan, MD, Laboratory of Molecular Biology, Division of Cancer Biology, National Cancer Institute, National Institutes of Health, 37/4E16, 37 Convent Dr MSC 4255, Bethesda, MD 20892.

![Fig. 2. Cytotoxicity and specificity of GM-CSF toxins on target cells. The colon lines SW403 (A) and LS174T (B) were incubated with GM-CSF–PE38KDEL and DT388–GM-CSF was incubated with the leukemia lines HL60 (C) and U937 (D) in the presence (•, ▴) or absence (○, ▵) of 5 μg/mL of GM-CSF. The cells were pulsed with [3H]-leucine and harvested, and the leucine incorporation was determined.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/1/10.1182_blood.v90.1.252/5/m_bl_0029f2.jpeg?Expires=1769272177&Signature=fSMdED2n0AvIASxRnivdGdcU6QzJamrculAApMQ1BwR6e4oj4mZWArKnzBqGM9m1XXZg40sMZBdnBoUa-SZQ-LNzejqpnFTqLlo~sdoOaeZvfto2-9f~081uwP-3FLPrqYOkc4Zlkg1-6lEeVx6h~Kmba2eG2VdD4fvLI9DtDJBkiC3JVNTkcdfkIkHNU519aNLXztZSKkh7pUeR3tD88b9ATCse1GbW70WKQsUUKq6zCFfHFNpx-d1070gMM-VQJBU8p-8DION02hKf69aJ87~XgfMQK1XFcaiA7zI0Jx79Yv16INGDSZUEOg1HAF2ladZ39fDcwJmzTGe3Rs85Wg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Scatchard plots of [125I]–GM-CSF binding to target cells. In (A), N87 cells (1.3 × 106/200 μL/well) were incubated with 0.15, 0.3, 0.6, 1.2, 2.4, and 4.8 nmol/L [125I]–GM-CSF (5.7 μCi/μg) in RPMI containing 0.1% bovine serum albumin and 0.2% NaN3 for 30 minutes, washed, and counted. HL60 cells (B) were assayed similarly at 2.2 × 106/well.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/1/10.1182_blood.v90.1.252/5/m_bl_0029f3.jpeg?Expires=1769272177&Signature=wHb0iai2YCPxlfkrfscmvBlSQN36PL1rnu3l8Oj7~tZqR3HRmgzZDjU1-3Py9du98e95FO4hpo3kqejWI8Uy-04ZUoPtnznX9ZQ6eDjAH6c3hT~bk9gaKOEHb9NI4kJob7E7Rhm9xYRWsuGZY0-uu2NtKEZ01qrfa28d94tWu~p68s69rjLnQnJkzMIMFFkl43Mjrlv5RATecJ7477wgcUenQz2kfi5~4X1-qcHxhGzlHcu9OJrNx~ZpoXEUJPTL~mu35iy2co3u-JyhE0MMga0P--setvtmgKw6fHRHK1Zud5cDxw3Y3wb7UcW-WSttM8hs8KWGRWf70U5I5rtTNQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. Cyotoxicity of recombinant toxins towards normal bone marrow cells. Normal marrow mononuclear cells (106/0.1 mL well) were incubated with DT388–GM-CSF (▵) or GM-CSF–PE38KDEL (○) for 60 hours and pulsed with [3H]-leucine. In (A), the cells were incubated in parallel with the negative control molecules PE38KDEL (•) or DT388-IL2 (▴). In (B), the cells were also incubated with DT388–GM-CSF (▪) or GM-CSF–PE38KDEL (□) in the presence of 20 ng/mL of GM-CSF.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/1/10.1182_blood.v90.1.252/5/m_bl_0029f4.jpeg?Expires=1769272177&Signature=axJ49yBIIkvi6CgGmz5sjFusOQfGJbfBtGzPLXatYLZR75LWgtUSCpZgJ8WvszNGRp4je0iLsCl-HU8xRa62USIJ2F8UKPxzw5IR2fBFLWM4viYRIuw8GYHLBO7lh3rYJp9HVuuOJcUm4453etwSjz~GwOqIv9aEb26H6ACL5dN54G7rJhGhXnrXNolYmFb11rBojG02EZ9HmuuCm~gVM7uJVYlIEU-Gl04EMmJrnMwQpya4z7i0CrbuWqVml~1RfNTZCUG0HvVC9tSKYRNQbjlfGbxF0gg1khO36ozBDE7-YGwf0hr0ZNuvk2xAjef6x5nlaLeXYw1rePpAn2CdIg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. Displacement analysis of recombinant toxins. In (A), U937 cells (4.5 × 106/well) were incubated with [125I]–GM-CSF and the indicated concentrations of GM-CSF (▪), GM-CSF–PE38KDEL (○), or DT388–GM-CSF (▵). In (B), HUT-102 cells (8 × 105/well) were incubated with [125I] — anti-TFR–IgG and the indicated concentrations of anti-TFR–IgG (▪), anti-TFR(Fv)–PE38KDEL (○), or DT388–anti-TFR(Fv) (▴). The cells were harvested and counted as in Fig 3.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/1/10.1182_blood.v90.1.252/5/m_bl_0029f5.jpeg?Expires=1769272177&Signature=uuBbXeHxy1gs~ndZKxb3F6sgyqZms5kMdI1sFlNeIS3wKEMxJCasWdMkEz2Gz7-qRYrAaNKcMAgykGAtO0ZUlwl5MUDIeq~Akcm5ZDqNzdv-7o5Z3RVntM2kuOZXhdSeFAcKUe03T-yosO-6P2UuvmwZpU7Glgg6ZTqYIW2kOtokpWf6YTulN4gUobBySfXKE48FM5cThTaKlnuhqkV1n3rfkDLmsoy6ETM4ZIk49urqHlXTEJA5SEqiSHIneQ07XTsva2hxfLuE6PRFCW40nTjIO2TJvFBf9yIzsRO8VkY7b-IG-eDT6qC~q9GM9EUU-FVFSISIqQPNV~rkwfgCaw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal