Abstract
Human CD23 is a 45-kD type II membrane glycoprotein, which functions as a low-affinity receptor for IgE and as a ligand for the CD21 and CD11b/CD11c differentiation antigens. CD23 is released from the surface of cells as soluble fragments, and a 25-kD species of soluble CD23 (sCD23) appears to act as a multifunctional cytokine. In this report, sCD23 is shown to sustain the growth of low cell density cultures of a human pre-B–acute lymphocytic leukemia cell line, SMS-SB: no other cytokine tested was able to induce this effect. Flow cytometric analysis indicates that sCD23 acts to prevent apoptosis of SMS-SB cells. SMS-SB cells cultured at low cell density possess low levels of bcl-2 protein. Addition of sCD23 to cells at low cell density maintained bcl-2 expression at levels equivalent to those observed in SMS-SB cells cultured at higher cell densities. No CD23 mRNA was found in SMS-SB cells, ruling out an autocrine function for CD23 in this cell line model. Although SMS-SB cells do not express the known receptors for CD23, namely CD21, CD11b-CD18, or CD11c-CD18, the cells specifically bind CD23-containing liposomes, but not glycophorin-containing liposomes. Binding of CD23-containing liposomes is inhibited by anti-CD23 but not by anti-CD21 or anti-CD11b/c monoclonal antibodies. The data show that sCD23 prevents apoptosis of the SMS-SB cell line by acting through a novel receptor.
THE HUMAN leukocyte differentiation antigen CD23 is a 45-kD type-II transmembrane glycoprotein, expressed on numerous hematopoietic lineage cells, which functions as a low-affinity receptor for IgE.1-4 The CD23 molecule is a member of the C-type (Ca2+-dependent) lectin family, and contains an α-helical coiled-coil stalk region between the extracellular lectin domain and the transmembrane region, which is believed to facilitate oligomerization of membrane-bound CD23 to trimers.5,6 Membrane-bound CD23 has roles in IgE binding,7,8 regulation of IgE synthesis,9 cell adhesion10,11 and antigen presentation.12 Two forms of 45-kD CD23 exist, CD23a and CD23b, which differ only in their cytosolic domains13; the former is constitutively expressed by B cells, and the latter is induced by exposure of the cells to interleukin-4 (IL-4).14 CD23 is usually expressed at the mature B-cell stage, mostly on activated cells, although some malignant pre-B cells from acute lymphoblastic leukemia patients can be induced to express CD23 after stimulation with IL-4.15
The membrane form of CD23 undergoes proteolysis, giving rise to soluble CD23 molecules of molecular weights 37 kD, 33 kD, 29 kD, 25 kD, and 16 kD. All fragments possess the lectin binding domain and, with the exception of the 16-kD fragment, retain the ability to bind IgE.1,16 A range of other cytokine activities have been attributed to soluble CD23 species, particularly to the 25-kD form referred to hereafter as sCD23.17 Thus, sCD23 has been shown to act as an autocrine growth factor in some Epstein-Barr virus (EBV)-transformed mature B-cell lines18 and as a differentiation factor for prothymocytes19 and also to prevent apoptosis of germinal centre B centrocytes,20 possibly via induction of bcl-2 expression.21 The putative pleiotropic cytokine activities of sCD23 are linked to a domain distinct from that involved in IgE binding.22 Elevated levels of sCD23 have been found in a range of clinical conditions such as rheumatoid arthritis,23 allergy,24 and B-chronic lymphocytic leukemia, in which sCD23 levels can be up to 500-fold higher than normal and the levels of sCD23 correlate with the clinical stage of the disease.25 Thus, there is strong circumstantial evidence that sCD23 is both biologically active and clinically important.
The reported receptors for CD23 are CD21,10 CD11b, and CD11c.26 CD21, a receptor for the C3d, C3g, and iC3b complement proteins, interferon-α and also EBV,27 is a 145-kD glycoprotein expressed on B lymphocytes, some T lymphocytes, follicular dendritic cells and pharyngeal epithelial cells.27,28 The CD23/CD21 interaction involves the C-type lectin activity of CD23,29 and CD21 and CD23 can therefore be regarded as a pair of adhesion molecules.10,11,30 Since CD21 is expressed by B and T cells and by follicular dendritic cells, the ability of sCD23 to prevent apoptosis in germinal centre cells20 has been postulated to occur via CD21. CD11b-CD18 and CD11c-CD18 are members of the β2 integrin family of adhesion proteins, and have recently been shown to act as receptors for CD23 on monocytic cells.26 Binding of CD23 to CD11b-CD18/CD11c-CD18 resulted in production of inflammatory cytokines including IL-1β, IL-6, and tumor necrosis factor-α, indicating a physiological role for CD23 in activation of monocytic cells.26
CD23 has potential roles in a range of cell systems, and may also act on hematopoietic precursors. In the case of B cells, the most likely receptor for CD23 action is CD21; however, CD21 is not expressed until the late pre-B–cell stage raising the possibility that other, as yet undefined, receptors for CD23 might exist within the B-cell precursor compartment. We have studied the effect of recombinant sCD23 on, and interaction of full-length CD23 with, a human pre-B–acute lymphocytic leukemia cell line, SMS-SB, which overexpresses the c-fos proto-oncogene.31 32 The data suggest that sCD23 prevents apoptosis of SMS-SB cells cultured at low cell density, and that sCD23 is interacting with the SMS-SB cells via a novel receptor.
MATERIALS AND METHODS
Materials
Protein-free hybridoma medium (PFHMII ), RPMI-1640, Dulbecco's minimal Essential medium (DMEM) and all tissue culture plasticware were obtained from Life Technologies Ltd (Paisley, Scotland). Fetal calf serum (FCS) was from Northumbria Biologicals (Durham, UK). A range of monoclonal antibodies were used, and these were obtained from DAKO Ltd (High Wycombe, UK); Serotec Ltd (Oxford, UK); Bradsure Biologicals (Loughborough, UK); and Pharmingen Ltd (Cambridge, UK). Radiochemicals, blotting media and photographic supplies were obtained from Amersham International plc (Little Chalfont, UK), and gel electrophoresis media were from Bio-Rad Laboratories Ltd (Watford, UK). RNAzol B was purchased from Biogenesis Ltd (Bournemouth, UK). POPC (1-palmitoyl-2-oleoyl-phosphatidyl choline) was purchased from Avanti Polar Lipids Inc (Alabaster, AL) and DiOC18 (3, 3′-dioctadecyloxacarbocyanine perchlorate) was obtained from Molecular Probes Ltd (Eugene, OR). Synthetic oligonucleotides were prepared in-house at the CRC Beatson Institute. Routine biochemicals were of the highest analytical grade available and were obtained from the Sigma Chemical Co (Poole, UK). B-cell lines used in these studies were from stocks held in individual laboratories, or were obtained from the European Collection of Animal Cell Cultures (Porton Down, UK). These lines were grown in either DMEM or RPMI-1640 media supplemented with 10%(vol/vol) FCS and 2 mmol/L fresh glutamine. Recombinant IL-4 was a generous gift from the Immunex Corp (Seattle, WA) and all other cytokines were purchased from British Biotechnology Ltd (Abingdon, UK).
Full-length 45-kD CD23 was expressed in baculovirus-infected Sf9 cells33 and has previously been shown to bind to CD23 receptors on B-cell lines10,29 and monocytic cells,26 and to have biological activity in human B-cell growth assays.33 Twenty-five kilodalton sCD23 was expressed either in baculovirus34 or in Escherichia coli as a polypeptide corresponding to amino acids 150-321 of CD239; the recombinant sCD23 has been shown to be active in rescue of germinal centre cells20 from apoptosis and induction of cytokine synthesis in monocytes.26 Both 45-kD full-length and 25-kD sCD23 species were affinity purified as previously described.33,34 The Rb55 rabbit antiserum was raised against the 25-kD sCD23 polypeptide (residues 150-321) expressed in E coli, and was employed as an affinity-purified IgG fraction.29 This reagent neutralizes the activity of CD23 in assays of human B-cell IgE production in vitro, and also blocks antigen-specific IgE responses in vivo when administered intravenously to rats.9
The SMS-SB Cell Line
The SMS-SB cell line is a human pre-B cell-like acute lymphocytic leukemia cell line, isolated from a female patient in 1981.31 The tumor presented as a solid paraspinal mass, with many features reminiscent of Abelson virus-induced pre-B–cell tumors in mice. Consistent with an acute lymphocytic leukemia (ALL) classification, SMS-SB cells contain cytoplasmic μ chains but are surface Ig negative, negative for CD10 (CALLA), and EBV-nuclear antigen expression, and do not display any of the chromosomal translocations commonly associated with lymphoproliferative tumors31; the cells show a degree of overexpression of the c-fos proto-oncogene.32 The cells adapted spontaneously to culture and are readily propagated in serum-free and protein-free media.
Cell Proliferation Assay
SMS-SB cells were propagated in PFHMII , harvested, and washed three times with PFHMII. The cells were then resuspended at 105 cells/mL in PFHMII and aliquotted at 1,000 or 2,500 cells/well in 96-well microtitre trays; the final volume of the cultures was 100 μL. Where appropriate, wells contained sCD23 or other cytokine at the doses indicated in the figure legends. Cultures were incubated at 37°C for 24 or 48 hours with 0.3 μCi/well [3H]-TdR being added to the wells for the final 6 hours of culture. Cultures were harvested onto glass fiber mats using an LKB Wallac cell harvester, and incorporation of radioactivity assessed by liquid scintillation spectrometry using an LKB Betaplate counter (LKB-Pharmacia Biotechnology, Milton Keynes, UK). All determinations were made in triplicate.
Preparation of Protein-Containing Fluorescent Liposomes
Preparation of fluorescent liposomes containing 45-kD CD23 or Glycophorin A was performed as described previously.10 29 Briefly, 10 μmol of POPC in chloroform were mixed with 50 nmol of DiO18 in dimethyl formamide to give a lipid ratio of 1/200, DiO18/POPC. After evaporation of solvents, by flushing with nitrogen, and further drying on a rotary evaporator, 50 μL of 800 mmol/L OGP (n-Octyl-β-D-glucopyranoside) in phosphate-buffered saline (PBS) was added to the tube and the solution vortexed until clear (15 to 30 minutes), whereupon the volume was adjusted to 200 μL with PBS (final concentration of OGP 200 mmol/L).
The protein to be incorporated into the fluorescent liposomes was dissolved in 50 mmol/L OGP in PBS (OGP/PBS) at 20 to 25 μg/mL. Approximately 15 μg of protein was added to the tube and OGP/PBS added to give a final volume of 800 μL. The mixture was vortexed for 10 seconds and then incubated on ice for 1 hour with occasional agitation. Finally, the liposomes were passed through a 0.22-μm filter and dialyzed against four × 1.2 L volumes of 20 mmol/L HEPES pH 7.0/140 mmol/L NaCl over a 48-hour period. After dialysis, the liposome stock solution was diluted 1/10 with liposome buffer (140 mmol/L NaCl, 2 mmol/L CaCl2 , 0.02% NaN3 , 0.5% bovine serum albumin [BSA], 20 mmol/L HEPES-KOH, pH 7.0) containing 1 mmol/L Nα-p-tosyl-L-lysine chloromethyl ketone and 10 mmol/L iodoacetamide and stored at 4°C in 5-mL aliquots.
Nucleic Acid Manipulations
Isolation of cellular RNA.Total RNA was isolated using RNAzol B, according to the manufacturer's instructions. Briefly, 106 to 107 cells were pelleted, washed in ice-cold PBS, lysed by addition of 0.2 to 2 mL RNAzol B. One-tenth volume of chloroform was added, the tube shaken vigorously for 15 seconds and left on ice for 5 minutes, and then centrifuged at 12,000g at 4°C for 15 minutes. The aqueous phase was removed, an equal volume of ice-cold isopropanol added and the RNA precipitated at −20°C overnight.
Reverse transcriptase-polymerase chain reaction (RT-PCR).For PCR amplification, a GeneAMP RNA PCR kit (Perkin Elmer Cetus, Beaconsfield, UK) was used per the manufacturer's instructions. Random hexamers (2.5 μmol/L) were used in the reverse transcription step plus 1 μg of the appropriate RNA, with incubations of 15 minutes at 42°C, 5 minutes at 99°C, and 5 minutes at 5°C. The amplification step employed Amplitaq DNA polymerase (Perkin Elmer Cetus) plus appropriate primers at 0.15 μmol/L. PCR amplification was performed in a Perkin Elmer Cetus DNA thermal cycler as follows: 2 minutes at 94°C for 1 cycle; 1 minute at each of 94°C, 55°C and 72°C for 35 successive cycles; 10 minutes at 72°C followed by storage at 5°C until retrieval.
Hybridization procedures.RT-PCR reaction products were electrophoresed on 0.8% (wt/vol) agarose/tris-borate-EDTA (TBE) nondenaturing gels and transferred to Hybond N+ nylon membranes under vacuum. The presence of specific products was achieved by probing with a 1.6-kb BamH1 fragment of the human CD23 cDNA clone pCDL SRα296CD23 labeled with α-[32P]–CTP by random priming, and visualization of hybridization by autoradiography.
Flow Cytometric Procedures
Cell phenotyping.For routine staining of cell surface markers, aliquots of 106 cells were harvested, washed twice in ice-cold PBS then incubated in 100-μL final volumes with fluorescein isothiocyanate (FITC)-conjugated MoAb to defined surface markers. After a 30 to 60 minute incubation on ice, the cells were washed twice in cold PBS, resuspended in 0.5 to 1.0 mL of the same buffer and propidium iodide added to a final concentration of 2 μg/mL immediately before data acquisition. Data were collected on a Becton Dickinson FACScan instrument (Abingdon, UK) and all histograms represent data from 104 live-gated events.
Liposome binding.SMS-SB cells were prepared as described above before addition of 50 μL of either CD23-containing fluorescent liposomes or glycophorin A-containing liposomes. After incubation at 4°C for 2 hours, the cells were washed three times with liposome buffer, resuspended in 100 μL of liposome buffer and analyzed. All manipulations were performed at 4°C before analysis. For antibody inhibition assays, either the liposomes or the cells (see individual experiments) were preincubated with the given antibody (at 10 μg/mL, unless otherwise stated) for 1 hour at 4°C before the liposomes were added to the cells.
Staining of bcl-2 in permeabilized cells.SMS-SB cells were cultured as previously with or without sCD23 and harvested for assay. For each stain, 106 cells were washed in 0.1% saponin/0.5% BSA in PBS (SBP buffer), incubated in SBP supplemented with 10% normal rabbit serum for 15 minutes, then washed twice in SBP. The cells were treated with 1 μg of anti-bcl-2 monoclonal antibody in a final volume of 100-μL SBP for 20 minutes on ice, before washing and addition of FITC antimouse IgG (γ-chain specific). After a 20-minute incubation and two washes, the cells were fixed in 2% paraformaldehyde/0.5% BSA in PBS for 15 minutes, washed twice with PBS, then suspended in a solution of 0.1% Triton X-100/0.1% sodium citrate/100 μg/mL propidium iodide before analysis on a Coulter Elite flow cytometer (Miami, FL).
Quantitation of apoptosis.Flow cytometric discrimination and quantitation of viable, early and late apoptotic and necrotic cells is based on their differential permeability and fluorescence following staining with the DNA-binding fluorochromes, Hoechst 33342 (Molecular Probes Inc), and propidium iodide.35
Cells were harvested and washed as noted above and, after instrument settings were adjusted for forward and side scatter using unstained cells, the test samples were analyzed. The cells were stained with freshly-prepared solutions of propidium iodide (20 μL of a 50 μg/mL solution in PBS) and Hoechst 33342 (100 μL of a 10 μg/mL solution) for 1 minute before collection of data from 10,000 cells on a dual laser Coulter Elite flow cytometer. In all experiments cell debris was excluded by electronic gating.
RESULTS
Twenty-Five Kilodalton sCD23 Sustains Proliferation of SMS-SB Cells
SMS-SB cells display a marked density-dependence for survival.36 Thus, plating the cells at high cell density (∼5 × 105/mL) gives cultures which grow rapidly, while plating at lower cell densities (≤5 × 104/mL) results in cultures in which no cells survive after 48 hours (data not shown). A range of cytokines were analyzed for their capacity to influence, either positively or negatively, the proliferation of SMS-SB cells cultured in PFHMII (Fig 1A). While the majority of cytokines tested had no effect upon SMS-SB cell proliferation, the data indicated that sCD23 appeared to facilitate the growth of SMS-SB cells plated at low cell density. Thus, the majority of the recombinant cytokines tested gave a stimulation index of approximately 1, while sCD23 gave a stimulation index of approximately 6; IL-5 stimulated SMS-SB to a lesser extent (stimulation index = 2.4). When SMS-SB cells were cultured at 2,500 cells/well for 48 hours in PFHMII in the presence of increasing concentrations of recombinant sCD23, an increase in tritiated-thymidine uptake with increasing sCD23 concentration was observed (Fig 1B). Consistent thymidine incorporation was observed at a sCD23 dose of approximately 10 ng/mL (400 pmol/L), which is in good agreement with concentrations necessary to elicit monocyte activation.26 The growth sustaining effect of sCD23 could be inhibited by the IgG fraction of a polyclonal anti-CD23 reagent (Rb55), but not by an equal amount of nonimmune rabbit IgG (Fig 1C). These data show that recombinant sCD23 sustains the growth of SMS-SB cells.
Effect of sCD23 on growth of SMS-SB cells. For each of the experiments illustrated in Fig 1, SMS-SB cells were harvested from log-phase cultures in PFMHII , washed, and seeded at low cell density (2.5 × 104 cells/mL) in the same medium; recombinant cytokines were included at 50 U/mL, and the final culture volume was 0.1 mL. The source of recombinant sCD23 used in these experiments was a Sf9 supernatant which was used at a final dilution of 1:100 in the cultures. The 24-hour cultures were pulsed with 0.3 μCi/well [3H]-TdR for 6 hours before harvest. All cultures were established in triplicate, and each experiment illustrated is representative of at least three independent repeats. (A) Presents the data for the effects of a range of individual cytokines, and stimulation indices were calculated according to the equation: Stimulation Index = Mean CPM incorporated in presence of cytokine / Mean cpm incorporated in absence of cytokine. (B) Presents a dose response curve using affinity-purified recombinant sCD23 produced in E coli.9 (C) Shows the effect of inclusion of 40 μg of the IgG fractions of RB55 anti-CD23 or nonimmune rabbit IgG on proliferation of SMS-SB cells cultured for 48 hours in the presence, or absence, of 100 ng/mL sCD23.
Effect of sCD23 on growth of SMS-SB cells. For each of the experiments illustrated in Fig 1, SMS-SB cells were harvested from log-phase cultures in PFMHII , washed, and seeded at low cell density (2.5 × 104 cells/mL) in the same medium; recombinant cytokines were included at 50 U/mL, and the final culture volume was 0.1 mL. The source of recombinant sCD23 used in these experiments was a Sf9 supernatant which was used at a final dilution of 1:100 in the cultures. The 24-hour cultures were pulsed with 0.3 μCi/well [3H]-TdR for 6 hours before harvest. All cultures were established in triplicate, and each experiment illustrated is representative of at least three independent repeats. (A) Presents the data for the effects of a range of individual cytokines, and stimulation indices were calculated according to the equation: Stimulation Index = Mean CPM incorporated in presence of cytokine / Mean cpm incorporated in absence of cytokine. (B) Presents a dose response curve using affinity-purified recombinant sCD23 produced in E coli.9 (C) Shows the effect of inclusion of 40 μg of the IgG fractions of RB55 anti-CD23 or nonimmune rabbit IgG on proliferation of SMS-SB cells cultured for 48 hours in the presence, or absence, of 100 ng/mL sCD23.
sCD23 Inhibits Apoptosis in Low Cell Density Cultures of SMS-SB Cells
To determine if SMS-SB cells were dying by apoptosis and that sCD23 acted to prevent apoptosis, cells were cultured at normal (5 × 105 cells/mL) and low cell density (2.5 × 104 cells per mL) in PFHMII for 24 hours in the presence and absence of sCD23 and examined by light microscopy (Fig 2). Cells cultured at normal cell density showed few apoptotic characteristics (Fig 2A) whereas cells cultured at low cell density (Fig 2B) exhibited a more spread morphology and became very adherent to the tissue culture flask; the cell membranes were blebbed and the cytoplasm was vacuolated, features which are characteristic of apoptotic cells (compare Fig 2A and B). Addition of sCD23 generated cells with a morphology indistinguishable from that of normal cell density cultures; fewer apoptotic cells were evident as the concentration of sCD23 was increased (Fig 2C and D).
Photomicrographs of SMS-SB cells.Cultures of SMS-SB cells in PFHMII were established at normal cell density (<105 cells/mL; A), or at low cell density (2.5 × 104 cells/mL) in the absence (B) or presence of 10 ng/mL (C) or 100 ng/mL sCD23 (D). After 24 hours of culture, the cells were inspected by bright field light microscopy under a 40× magnification.
Photomicrographs of SMS-SB cells.Cultures of SMS-SB cells in PFHMII were established at normal cell density (<105 cells/mL; A), or at low cell density (2.5 × 104 cells/mL) in the absence (B) or presence of 10 ng/mL (C) or 100 ng/mL sCD23 (D). After 24 hours of culture, the cells were inspected by bright field light microscopy under a 40× magnification.
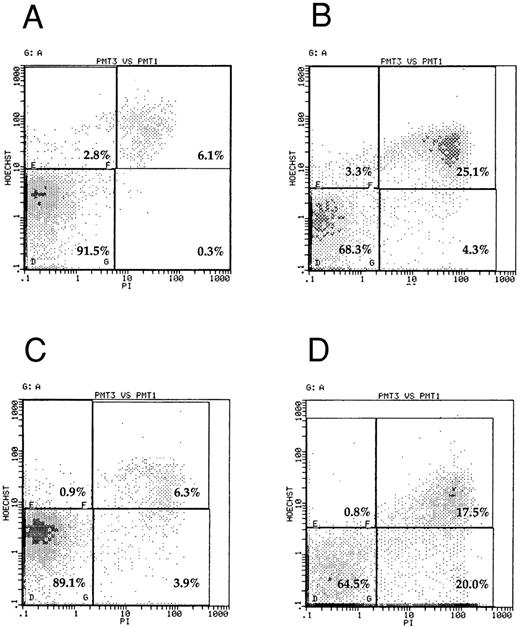
To confirm that SMS-SB cells in low cell density cultures were undergoing apoptosis, two-laser flow cytometric analysis of SMS-SB cells harvested after 24 hours of culture at normal or at low cell density either alone or in the presence of sCD23 or IL-4 was performed (Fig 3). The cells were stained with propidium iodide and Hoechst 33342 and, after gating on the basis of scatter parameters, analyzed by two-laser flow cytometry. In cultures propagated at normal cell density (Fig 3A), approximately 90% of the cells were routinely located in the lower left quadrant of the dot-plots, indicating viable cells (91.5% in this example); few dead/necrotic cells were evident (lower right quadrant). In normal cell density cultures, some 6% of cells scored as late apoptotic (upper right quadrant). These values contrast sharply with the values for cells cultured at low cell density (Fig 3B) where the number of viable cells was 68.3% and the number of late apoptotic cells was approximately 25%. Inclusion of IL-4 in these cultures (Fig 3D) resulted in a small decrease in late apoptotic cells, but the total number of viable cells in the cultures remained comparatively low at approximately 65%, and there was a sizeable population of dead cells in cultures containing IL-4. However, the presence of sCD23 in the low cell density cultures (Fig 3C) resulted in restoration of the number of viable cells in the culture to approximately 90% (89.1% in this example), and a reduction of late apoptotic cells to 6.3% of the culture, values which are essentially identical to those observed for normal cell density cultures. The capacity of sCD23 to block apoptosis in low cell density cultures was concentration dependent; thus 10 ng/mL sCD23 resulted in approximately 12% of the cells scoring as late apoptotic compared with 6% when 100 ng/mL sCD23 was included in the cultures (data not illustrated). The data indicate that sCD23 prevents entry of SMS-SB cells into apoptosis under low cell density culture conditions.
Flow cytometric analysis of apoptosis in SMS-SB cells. Cultures of SMS-SB cells in PFHMII were established as described in the legend to Fig 2. After 24 hours of culture, the cells were harvested, stained simultaneously with propidium iodide and Hoechst 33342, and after gating on forward and side scatter parameters, analyzed by two-laser cytometry.35 (A) And (B), respectively, present the data for SMS-SB cells obtained from normal and low cell density cultures, and (C) and (D) illustrate the effect of either 100 ng/mL sCD23 or 20 U/mL IL-4, respectively, on the extent of apoptosis in the cultures. On the individual panels, the percentage figures shown on the lower left quadrant represent viable cells and those on the lower right represent dead cells. Values in the upper left quadrant refer to early apoptotic cells while those in the upper right represent late apoptotic cells. The experiment shown is representative of four independent repeats.
Flow cytometric analysis of apoptosis in SMS-SB cells. Cultures of SMS-SB cells in PFHMII were established as described in the legend to Fig 2. After 24 hours of culture, the cells were harvested, stained simultaneously with propidium iodide and Hoechst 33342, and after gating on forward and side scatter parameters, analyzed by two-laser cytometry.35 (A) And (B), respectively, present the data for SMS-SB cells obtained from normal and low cell density cultures, and (C) and (D) illustrate the effect of either 100 ng/mL sCD23 or 20 U/mL IL-4, respectively, on the extent of apoptosis in the cultures. On the individual panels, the percentage figures shown on the lower left quadrant represent viable cells and those on the lower right represent dead cells. Values in the upper left quadrant refer to early apoptotic cells while those in the upper right represent late apoptotic cells. The experiment shown is representative of four independent repeats.
sCD23 Influences bcl-2 Expression in SMS-SB Cells
Expression of bcl-2 in SMS-SB cells was determined by immunostaining of bcl-2 protein and propidium iodide staining of DNA content of permeabilzed SMS-SB cells following a variety of culture conditions (Fig 4). Flow cytometric analysis of normal cell density cultures revealed a strong G1 peak (Fig 4A), with a significant proportion of the cells in S or G2/M phases, and greater than 97% of the cells scoring positive for bcl-2 expression. In contrast, cells from the low cell density culture (Fig 4B) were mainly in a pre-G1 peak with weak staining for bcl-2. However, in the presence of sCD23 (Fig 4C), there was a restoration of the cell cycle profile characteristic of asynchronously growing cells, and strong bcl-2 staining was observed in all phases of the cell cycle; in the example shown, 92.2% of the cells were bcl-2+. These results suggest that sCD23 treatment of low cell density cultures of SMS-SB cells may sustain bcl-2 expression and prevent apoptosis.
Flow cytometric analysis of bcl-2 expression in SMS-SB cells. SMS-SB cells were cultured as described in the legend to Fig 3, harvested and washed. The cells were gently permeabilized in 0.1% (wt/vol) saponin-containing buffer and treated with monoclonal anti-bcl-2 antibody. After incubation for 20 minutes on ice, the cells were washed and an aliquot of FITC-anti-mouse IgG added to visualize the primary MoAb. After further incubation and washing, the cells were treated with 100 μg/mL propidium iodide and immediately analyzed by flow cytometry. In the dot plots illustrated, the X-axis illustrates propidium iodide staining in linear units of fluorescence, while the Y-axis represents bcl-2 staining on a logarithmic scale. The experiment shown is representative of three repeats. (A) Normal cell density. (B) Low cell density. (C) Low cell density + 100 ng/mL sCD23.
Flow cytometric analysis of bcl-2 expression in SMS-SB cells. SMS-SB cells were cultured as described in the legend to Fig 3, harvested and washed. The cells were gently permeabilized in 0.1% (wt/vol) saponin-containing buffer and treated with monoclonal anti-bcl-2 antibody. After incubation for 20 minutes on ice, the cells were washed and an aliquot of FITC-anti-mouse IgG added to visualize the primary MoAb. After further incubation and washing, the cells were treated with 100 μg/mL propidium iodide and immediately analyzed by flow cytometry. In the dot plots illustrated, the X-axis illustrates propidium iodide staining in linear units of fluorescence, while the Y-axis represents bcl-2 staining on a logarithmic scale. The experiment shown is representative of three repeats. (A) Normal cell density. (B) Low cell density. (C) Low cell density + 100 ng/mL sCD23.
SMS-SB Cells Do Not Express CD23
The expression of CD23 by SMS-SB cells was examined to address the possibility that CD23 was acting on SMS-SB cells in an autocrine manner, as has been shown for some EBV-transformed B-cell lines.18 Flow cytometric analysis showed that SMS-SB cells had no CD23 on their surface but SKW cells (a mature human B-cell line) had readily detectable levels of CD23 (data not shown). RT-PCR was performed, with CD23-specific primers, on RNA isolated from SMS-SB cells and from SKW, Ramos, and Jijoye B-cell lines and from a keratinocyte cell line as a negative control. After 35 cycles, products of the reaction were electrophoresed and detected by Southern blotting (Fig 5). Ramos and SKW both generated strong signals of 256 bp. No signals were detected using SMS-SB, Jijoye, or keratinocyte RNA and there was no product from a reaction performed with no template RNA. Transcripts of c-fos were readily detectable in SMS-SB cells propagated under either protein-free or serum-supplemented culture conditions (data not shown). Figure 5 also indicates that SMS-SB cells do not express any RNA for CD23 when cultured in protein free media or in medium supplemented with 10% serum. These data negate the hypothesis that SMS-SB cells express CD23 and utilize this cytokine in an autocrine loop to prevent apoptosis.
PCR analysis of CD23 expression in SMS-SB and other B-lymphoid cell lines. Total cellular RNA was prepared from a range of B-lymphoid and keratinocyte cell lines, and from SMS-SB cultured either in PFHMII or in serum-containing medium. Total cellular RNA was used as template for RT-PCR using the primers indicated and the products were separated by agarose gel electrophoresis and probed with a [32P]-labeled insert from the plasmid pCDL SRα296CD23. Hybridization was visualized by autoradiography.
PCR analysis of CD23 expression in SMS-SB and other B-lymphoid cell lines. Total cellular RNA was prepared from a range of B-lymphoid and keratinocyte cell lines, and from SMS-SB cultured either in PFHMII or in serum-containing medium. Total cellular RNA was used as template for RT-PCR using the primers indicated and the products were separated by agarose gel electrophoresis and probed with a [32P]-labeled insert from the plasmid pCDL SRα296CD23. Hybridization was visualized by autoradiography.
SMS-SB Cells Do Not Express Known CD23 Receptors
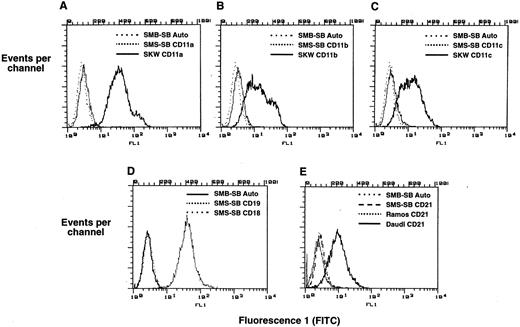
The data of Figs 1-4 show that sCD23 exerts a biological effect on SMS-SB cells raising the question of which CD23 receptor mediates this biological effect. The expression of known CD23 receptors by SMS-SB cells was assessed by flow cytometry as shown in Fig 6. SMS-SB cells and SKW cells were stained with FITC-labeled MoAb to each of CD11a, CD11b, and CD11c, and the data of Fig 6, A through C, respectively, clearly illustrate that while SKW cells express high levels of all three CD11 species, the level of staining of SMS-SB cells was superimposable on the autofluorescence profile for SMS-SB cells. Thus, SMS-SB cells do not express any of the CD11a, CD11b, or CD11c markers on their surface. Staining with FITC-anti-CD18 (Fig 6D) illustrates that SMS-SB cells also lack this component of the β2 integrin complex. Figure 6E shows a flow cytometric analysis of CD21 expression by SMS-SB cells in comparison with controls of the mature B-cell lines, Daudi, and Ramos. Daudi cells were strongly positive for CD21 expression whereas Ramos were only weakly positive. SMS-SB cells were negative for expression of CD21 at their surface. Neither SMS-SB cell cultures propagated in protein-free media nor those cultured in serum-supplemented media had detectable levels of Poly A+ RNA for CD21 and no CD21 transcripts were detected by RT-PCR (data not illustrated). The data are consistent with the hypothesis that SMS-SB cells possess a receptor for CD23 distinct from CD21 and the CD11 family members.
Expression of CD21 and CD11 antigens by SMS-SB cells. Aliquots of SMS-SB and other B-lymphoid cell lines were harvested, washed in PBS, and stained with FITC-conjugated MoAbs specific for the CD11a, CD11b, CD11c, CD18, CD19, and CD21 antigens. After a 30- to 60-minute incubation, the cells were washed, and analyzed by flow cytometry; nonviable cells were excluded from acquisition by propidium iodide counter-staining, and each graph represents the data from 10,000 live-gated events. Line styles are indicated on the individual panels, and “auto” refers to autofluorescence of unstained cells. (A) CD11a expression on SKW and SMS-SB cells. (B) CD11b expression on SKW and SMS-SB cells. (C) CD11c expression on SKW and SMS-SB cells. (D) CD18 and CD19 expression on SMS-SB cells. (E) Expression of CD21 on Daudi, Ramos, and SMS-SB cells.
Expression of CD21 and CD11 antigens by SMS-SB cells. Aliquots of SMS-SB and other B-lymphoid cell lines were harvested, washed in PBS, and stained with FITC-conjugated MoAbs specific for the CD11a, CD11b, CD11c, CD18, CD19, and CD21 antigens. After a 30- to 60-minute incubation, the cells were washed, and analyzed by flow cytometry; nonviable cells were excluded from acquisition by propidium iodide counter-staining, and each graph represents the data from 10,000 live-gated events. Line styles are indicated on the individual panels, and “auto” refers to autofluorescence of unstained cells. (A) CD11a expression on SKW and SMS-SB cells. (B) CD11b expression on SKW and SMS-SB cells. (C) CD11c expression on SKW and SMS-SB cells. (D) CD18 and CD19 expression on SMS-SB cells. (E) Expression of CD21 on Daudi, Ramos, and SMS-SB cells.
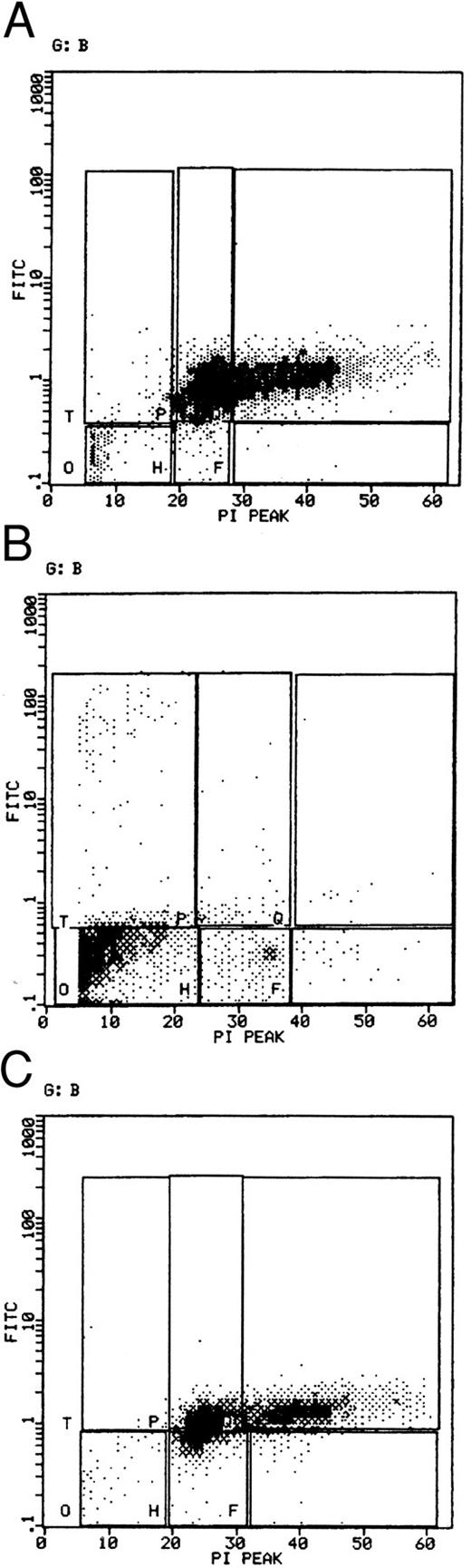
Specific Binding of CD23-Containing Liposomes to SMS-SB Cells
Since SMS-SB cells do not express any of the currently characterized receptors for CD23 (CD21, CD11b, and CD11c), a formal demonstration of CD23 binding to SMS-SB cells was sought. Liposomes containing full-length CD23 were prepared, incubated with SMS-SB cells and binding to the cells quantitated by flow cytometry. Figure 7 shows binding of CD23-containing liposomes to both SMS-SB (A) and RPMI-8226 cells (B), but not to Raji cells (C); glycophorin-containing liposomes did not bind to any of these cell lines. Liposome binding to SMS-SB cells via the novel CD23 receptor was of a similar level to that observed for RPMI-8226 cells that bind CD23 via CD21. Binding of CD23-containing liposomes to both SMS-SB and RPMI-8866 cells was prevented by inclusion of EDTA in the binding reaction medium (data not shown), which suggests that the C-type lectin activity of CD23 is required for binding to the novel receptor on SMS-SB cells.
Quantitation and specificity of liposome binding to SMS-SB and other B-lymphoid cell lines. Aliquots of SMS-SB (A), RPMI-8226 (B), and Raji cells (C) were mixed with either CD23-containing (solid line) or glycophorin-containing (broken line) fluorescent liposomes, washed and analyzed by flow cytometry for liposome binding. (D) Illustrates the binding data for SMS-SB and RPMI-8226 cells exposed to CD23-containing liposomes, which had been preincubated for 1 hour with 10 μg of EBVCS4 or MoAb25 anti-CD23 MoAbs before addition to the cells; the autofluorescence pattern of each cell is illustrated as a broken line.
Quantitation and specificity of liposome binding to SMS-SB and other B-lymphoid cell lines. Aliquots of SMS-SB (A), RPMI-8226 (B), and Raji cells (C) were mixed with either CD23-containing (solid line) or glycophorin-containing (broken line) fluorescent liposomes, washed and analyzed by flow cytometry for liposome binding. (D) Illustrates the binding data for SMS-SB and RPMI-8226 cells exposed to CD23-containing liposomes, which had been preincubated for 1 hour with 10 μg of EBVCS4 or MoAb25 anti-CD23 MoAbs before addition to the cells; the autofluorescence pattern of each cell is illustrated as a broken line.
Preincubation of the liposomes for 1 hour with the anti-CD23 MoAbs EBVCS4 or MoAb25 inhibited binding of CD23-containing liposomes to SMS-SB cells (Fig 7D). Numerous other antibodies, including a panel of anti-CD21 MoAbs, anti-CD11a, anti-CD11b, anti-CD11c, and anti-CD18 MoAbs were tested for their ability to prevent CD23-liposome binding to SMS-SB cells. All of the anti-CD21 antibodies employed failed to inhibit liposome binding. The rationale for testing anti-β2 integrin MoAbs was that failure to detect high levels by flow cytometry would not preclude binding activity if these molecules were expressed at a very low level; the data of Table 1 indicate that no inhibition of liposome binding was observed with MoAbs specific for CD11a, CD11b, CD11c, and CD18. Finally, MoAbs to a range of other markers expressed on SMS-SB cells failed to block liposome binding (Table 1). Only the EBVCS4 and MoAb25 anti-CD23 antibodies were able to block binding of the CD23-containing liposomes to the cells. The liposome binding and inhibition studies are consistent with the hypothesis that SMS-SB cells possess a novel receptor for CD23.
Antibody Inhibition of CD23 Liposome Binding
| MoAb Added . | Inhibition of CD23-Liposome Binding . |
|---|---|
| None | − |
| CD23* | + |
| CD5† | − |
| CD11a | − |
| CD11b | − |
| CD11c | − |
| CD18 | − |
| CD19† | − |
| CD21‡ | − |
| CD22 | − |
| CD54† | − |
| MoAb Added . | Inhibition of CD23-Liposome Binding . |
|---|---|
| None | − |
| CD23* | + |
| CD5† | − |
| CD11a | − |
| CD11b | − |
| CD11c | − |
| CD18 | − |
| CD19† | − |
| CD21‡ | − |
| CD22 | − |
| CD54† | − |
SMS-SB cells were harvested, washed, and preincubated with antibodies of the specificities noted in the Table for 1 hour at 4°C. After further washing to remove unbound antibody, CD23-containing liposomes were added and binding assessed by flow cytometry. Binding was scored in relation to the data presented in Fig 7; eg, − represents maximal binding of liposomes to SMS-SB cells (no inhibition) and + is equivalent to the inhibition noted in Fig 7 with EBVCS4.
Liposomes were preincubated with anti-CD23 antibody rather than SMS-SB cells; EBVCS4 and MoAb 254 were used, and Rb55 also partially inhibited liposome binding.
The antigen is detectable by direct FITC-MoAb staining of SMS-SB cells.
The following anti-CD21 MoAbs were tested: BU33 (The Binding Site, Birmingham, UK), THB5 (GBRI), MCA644 (Serotec), CR2 (Becton Dickinson), and IOB1a (Immunotech, Marseille, France).
DISCUSSION
The principal findings of this report are that addition of soluble CD23 to a human pre-B–ALL cell line, SMS-SB, results in rescue of the cells from apoptosis, possibly by influencing expression of bcl-2 in the cells. The cells do not express the known receptors for CD23, namely CD21, CD11b-CD18, and CD11c-CD18, but bind CD23-containing liposomes specifically, suggesting the existence of a novel CD23-binding structure on SMS-SB cells. In this model, sCD23 is not acting in an autocrine fashion as SMS-SB cells do not express CD23 RNA.
An overtly mitogenic mode of action of sCD23 on SMS-SB cells appears unlikely. Based on paradigms in the mature B-cell compartment, and parallels with early T-cell and myeloid development, a working hypothesis that the novel sCD23 receptor on SMS-SB cells acts to restrain apoptosis of the cells is attractive. In the majority of reports on sCD23 activity, including the thymocyte19 and centrocyte systems,20 the effects of sCD23 are only observed after pretreatment of the cells with IL-1. In these systems, IL-1 may preactivate the cells, priming them to respond to a signal from sCD23; this may occur via induction or increase in number or affinity of CD23 receptors at the cell surface. In the germinal centre model, the requirement for IL-1 can be overcome by anti-CD21 MoAbs, a subset of which are capable of preventing centrocyte apoptosis.37 However, no endogenous IL-1α or IL-1β activity is present in SMS-SB cell supernatants, and exogenous cytokine, either alone or in combination with sCD23, has any effect on proliferation or prevention of apoptosis in SMS-SB cells (data not illustrated); sCD23 alone is sufficient to completely prevent apoptosis in SMS-SB cells cultured at low cell density. The transformed state of SMS-SB cells may explain the absence of a requirement for IL-1, or other form of cellular preactivation, for the expression of sCD23 biological activity.
The data of this report illustrate the existence of a receptor for CD23 distinct from CD21 and CD11b-CD18 and CD11c-CD18. At present no detailed biochemical data are available for this receptor. However, it is likely that many of the features of sCD23 binding to other receptors will be found in this system. Clearly, the oligomeric state of the CD23 molecule has profound implications for its ability to bind to receptors, and the low affinity interactions mediated via the lectin domain of the CD23 molecules appear to be enhanced by oligomerization. Protein chemical cross-linking studies of membrane-bound and soluble CD23 have suggested that the α-helical coiled coil stalk region of the molecule facilitates trimerization of membrane-associated CD23.5,6 Such trimers would have three sites for ligand binding, which could be occupied by any combination of IgE, CD21, or CD11b-CD18/CD11c-CD18. Twenty-five kilodalton sCD23, despite lacking the stalk region,6 forms hexamers in solution which are thought to be a dimer of trimers, and to self-associate noncovalently via motifs on the ends of the lectin domains which are sterically hindered in the intact form. This would give rise to a molecule with six sites for ligand contact in which the intrinsically low affinity of binding of any one site would be overcome by the high avidity of a hexameric complex.
Treatment of low cell density cultures of SMS-SB cells with sCD23 results in inhibition of apoptosis, possibly via effects on expression of bcl-2. Although our data are based on investigations of a single pre-B cell-like cell line, it is tempting to speculate that CD23 may have a role in normal and neoplastic B lymphopoiesis; this notion is supported by the observation that the high levels of sCD23 observed in certain B-chronic lymphocytic leukemias are related to the clinical stage of the disease.38 Moreover, bone marrow stromal cells can be induced to express CD23,39 raising the possibility that CD23 could act in adhesion or soluble modes, or both, in delivery of signals which might influence B lymphopoiesis in the bone marrow microenvironment. If CD23 has a role in prevention of apoptosis in B-cell precursors, then CD23 might act independently to provide a rescue signal or, perhaps more likely, could act in synergy with other cytokines and adhesion strucutres to modulate the fate of B-lymphoid progenitors. These issues will be resolved only when detailed knowledge of the CD23 receptor present on SMS-SB cells is available and the pattern of its expression in normal B-cell progenitors can be assessed.
The SMS-SB cell line provides a very useful model for the study of the novel sCD23/CD23 receptor whose existence is suggested by the data presented in this report; the cells will be particularly useful in the analysis of mechanisms of CD23 action in the absence of other known receptors. The data show that ligation of the novel CD23 receptor prevents apoptosis of SMS-SB cells and, once the nature of the receptor is defined, it will be interesting to examine its expression and possible functions at different stages in the normal and neoplastic development of B lymphocytes and other hematopoietic lineages.
ACKNOWLEDGMENT
We thank Dr G. Webster (University of Glasgow) for expert technical assistance with flow cytometry.
L.J.W. is a Biotechnology and Biological Sciences Research Council (BBSRC) postgraduate scholar.
Address reprint requests to William Cushley, PhD, Division of Biochemistry & Molecular Biology, Institute of Biomedical & Life Sciences, University of Glasgow, Glasgow G12 8QQ, Scotland, UK.

![Fig. 1. Effect of sCD23 on growth of SMS-SB cells. For each of the experiments illustrated in Fig 1, SMS-SB cells were harvested from log-phase cultures in PFMHII , washed, and seeded at low cell density (2.5 × 104 cells/mL) in the same medium; recombinant cytokines were included at 50 U/mL, and the final culture volume was 0.1 mL. The source of recombinant sCD23 used in these experiments was a Sf9 supernatant which was used at a final dilution of 1:100 in the cultures. The 24-hour cultures were pulsed with 0.3 μCi/well [3H]-TdR for 6 hours before harvest. All cultures were established in triplicate, and each experiment illustrated is representative of at least three independent repeats. (A) Presents the data for the effects of a range of individual cytokines, and stimulation indices were calculated according to the equation: Stimulation Index = Mean CPM incorporated in presence of cytokine / Mean cpm incorporated in absence of cytokine. (B) Presents a dose response curve using affinity-purified recombinant sCD23 produced in E coli.9 (C) Shows the effect of inclusion of 40 μg of the IgG fractions of RB55 anti-CD23 or nonimmune rabbit IgG on proliferation of SMS-SB cells cultured for 48 hours in the presence, or absence, of 100 ng/mL sCD23.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/1/10.1182_blood.v90.1.234/5/m_bl_0013f1.jpeg?Expires=1770441380&Signature=lRdMbMERSxUrBHqv7QZx9qt7xql8L~A-ijhExyKzoNO6Jg1B1O-BJ242I1tD8EGu6M5k7bgLa1uFhwga9rFGEan3Se3D8KV-4DW1s4GLhpZEU8LA1aunaoURgryzYxGw-DrCW~NnqChaPITGhx8V02UQRZ6Fj8UodfeNnaFPM7g65EuO3oPIr6ui8wd9-iYfRuJSxzWltuh0DCgFCSjlxnglkOeCfnCE7YeH1P3i78NoQ-qZn-Tn~qVnLDW5UNBOX2WcTolTLSSrKXdvo06SrIsiiQnHpwAv~I-YPsIkgOeHhFTse3e2w9pgub--crVUV9-qczWKd1VLJhX1VgkyXA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 5. PCR analysis of CD23 expression in SMS-SB and other B-lymphoid cell lines. Total cellular RNA was prepared from a range of B-lymphoid and keratinocyte cell lines, and from SMS-SB cultured either in PFHMII or in serum-containing medium. Total cellular RNA was used as template for RT-PCR using the primers indicated and the products were separated by agarose gel electrophoresis and probed with a [32P]-labeled insert from the plasmid pCDL SRα296CD23. Hybridization was visualized by autoradiography.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/1/10.1182_blood.v90.1.234/5/m_bl_0013f5.jpeg?Expires=1770441380&Signature=i2M1a~HDCRBwjb6WHH5~kMcFSBi8q8YOua9N8uCiXbnnHEEZvjlknxpJVqWFvGNNeqtsi8I9EYNzls0jtdLtz1Uf9t1Sx7tplaT8Cu8RpWRMeBS9H7MnNMwVs1n2EFM9ycswNeM-FvaqbkEDUaWQfH6r4vleJ5aHJWOb5CktBbure~EAgtu9NqssEESEaKRFcbWu1wJFAHPTaBuCeJpqNea-ntqNtw5A7g0RC3xYehOIwslMC-WmKbE7ntpnXp42U0sHhzR~owGuPGBY92E9H~Zqukh1xR9XWZn2fUF4moULwFQrVCMCUnL9LP3MGrvoM1KlwmESWFi~riXC4THlzA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal