Abstract
Chemokines are a family of related proteins that regulate leukocyte infiltration into inflamed tissue. Some chemokines such as MIP-1 α also inhibit hematopoietic progenitor cell proliferation. Recently, three chemokines, MIP-1 α, MIP-1 β, and RANTES, have been found to significantly decrease human immunodeficiency virus production from infected T cells. We report here the cloning and characterization of a novel human chemokine termed Exodus for its chemotactic properties. This novel chemokine is distantly related to other chemokines (28% homology with MIP-1 α) and shares several biological activities. Exodus is expressed preferentially in lymphocytes and monocytes, and its expression is markedly upregulated by mediators of inflammation such as tumor necrosis factor or lipopolysaccharide. Purified synthetic Exodus was found to inhibit proliferation of myeloid progenitors in colony formation assays. Exodus also stimulated chemotaxis of peripheral blood mononuclear cells. The sequence homology, expression, and biological activity indicate that Exodus represents a novel divergent β-chemokine.
CHEMOKINES are a family of related proteins that regulate leukocyte migration and activation in the face of inflammatory stimulus.1-5 They are secreted by activated leukocytes themselves, by endothelial and epithelial cells, and by some types of malignancies.1-5
Chemokines consist of 8 to 12 kD, basic, heparin-binding proteins that have four cysteines conserved among all family members. The chemokine family can be subdivided depending on where the first two cysteines are located. If they are separated by a single amino acid, they are classified as the α-family (also called C-X-C). If they are adjacent to each other they are classified in the β family (also called C-C). α Members are located in a gene cluster at human chromosome 4q12-21, and β members in a cluster at human chromosome 17q11-32.6-8
Leukocytes respond to chemokines through receptors that are seven membrane spanning G-protein–coupled receptors.9-11 Two receptors have been identified for C-X-C chemokines, and five C-C receptors have been characterized. These receptors may play a role in human disease. For example, it appears that a red blood cell chemokine receptor is the critical malaria receptor.12
Chemokines have a wide range of biological activity. They initially were described by their ability to stimulate leukocyte chemotaxis and activation.5 Because of these proinflammatory activities, they play a role in a wide variety of diseases that have inflammatory tissue destruction, such as rheumatoid arthritis, myocardial infarction, and adult respiratory distress syndrome.13-18
β-Chemokines can be further subdivided according to structural homologies and similar activities. MIP-1 α, MIP-1 β, and RANTES have closer homology and range of biological activities than the other members of the family.1,4,5 Another subtype within the β-chemokine family is called monocyte chemotactic proteins (MCP) according to their ability to preferentially stimulate monocytes to migrate and respond to inflammatory stimuli.19 These proteins are also more structurally similar to each other than other members of the family.
Some chemokines, especially members of the β family such as MIP-1 α, have the ability to decrease the proliferative status of myeloid progenitors.20-23 This effect occurs in primitive hematopoietic progenitors, perhaps even at the stem cell level. This proliferative inhibition may protect hematopoietic progenitors from cell cycle-specific chemotherapeutic cytotoxicity.23
Recently, there have been several reports that the beta chemokines MIP-1 α, MIP-1 β, and RANTES inhibit human immunodeficiency virus production in newly infected peripheral blood mononuclear cells (PBMCs).24-26 The HIV coreceptors (companions to CD4) are chemokine receptors, suggesting a mechanism for the inhibition of HIV production by some β-chemokines.27-34
We have isolated and characterized a novel but divergent member of the β-chemokine family, called Exodus for its chemotactic properties. Exodus is expressed preferentially in lymphocytes, and is markedly upregulated by inflammatory stimuli. Exodus exhibits similar activities to other β-chemokines.
MATERIALS AND METHODS
Cloning.Messenger RNA was prepared from dissected human pancreatic islet cells. A unidirectional cDNA library with inserts greater than 1,000 nucleotides was constructed in the vector lambda ZAPII using the uni-ZAPTMXR system (Stratagene, La Jolla, CA). The library was en masse excised from the vector in pBluescript SK- according to the manufacturer's instructions. More than 1,000 plasmid clones were randomly selected and single pass dideoxy-sequenced from either the 5′ and/or the 3′ ends using an ABI DNA sequencer (Applied Biosystems, Foster City, CA). The sequences of these expressed sequence tags (EST) has been deposited in GenBank.35
One of the sequenced clones had distant homology to the β-chemokine family when a consensus chemokine sequence was used to search against these pancreatic expressed sequences. This homology became much more closely aligned to the chemokine family when several sequencing errors from the original automated pass were corrected by manual dideoxy double-stranded sequencing. This EST clone, originally designated HBC2850, on complete sequencing was not found to be identical to any other known chemokine. This cDNA of 821 nucleotides contained the entire open reading frame of the gene (GenBank Accession number U64197). Based on the well-described chemotactic properties of the β-chemokine family, we termed this novel chemokine gene Exodus.
A human macrophage cDNA library was also screened with primers specific for HBC 2850, and a second Exodus cDNA isolated. This macrophage cDNA encoded the identical protein as the pancreatic cDNA, with the exception of an extra codon for alanine (GCA) at nucleotide position 120. This most likely represents an allelic polymorphism.
Cell culture.The monocytic cell line THP-1 was obtained from American Type Culture Collection (Rockville, MD). Cells were maintained in RPMI 1640 media supplemented with 10% fetal calf serum (FCS), 25 mmol/L HEPES, and antibiotics. For stimulation experiments, cells were cultured at a density of one million per mL in the presence of phorbol ester (PMA, Sigma, St Louis, MO). The immortalized human umbilical vein endothelial cell line I-HUVEC was obtained from Dr Jay Nelson (University of Oregon, Eugene), and cultured in RPMI 1640 supplemented with 10% FCS, 400 μg/mL G418 (Life Technologies, Grand Island, NY), 1 μ/mL heparin (Sigma), and 30 μg/mL endothelial cell growth factor (Collaborative Biomedical Products, Bedford, MA). Cells were grown to a confluency of 70% to 80%, then cultured in the presence or absence of 10 ng/mL tumor necrosis factor-α (TNF-α) for various periods of time. PBMCs were purified on Histopaque gradients (Sigma) and monocytes were isolated by plastic adherence. Monocytes were cultured for 6 days, with media replaced every 2 days, to allow for differentiation into macrophages. Cells were stimulated with 100 ng/mL lipopolysaccharide. (LPS, Sigma) for various time periods.
The cytokine-dependent primitive human myeloid leukemia cell line MO7E was cultured in RMPI 1640 plus 10% FCS and antibiotics. This media was supplemented with granulocyte-macrophage colony stimulating factor (GM-CSF, 100 U/mL) and Steel factor (SLF, also called stem cell factor, 50 ng/mL) for maximal log phase growth. These cytokines were obtained from Immunex Corp (Seattle, WA).
Expression analysis.The probe for Northern analysis was the cDNA containing the complete coding region of Exodus, isolated by agarose gel electrophoresis, and labeled with P32-dCTP and P32-dTTP (DuPont-NEN, Boston, MA) by random priming according to the manufacturer's instructions (BMB, Indianapolis, IN). RNA was isolated from cell lines and cultured monocytes using RNA STAT-60 (Tel-Test B Inc, Friendswood, TX) according to the manufacturer's instructions. Total RNA (20 μg) was fractionated on 0.8% formaldehyde agarose gels, transferred to nitrocellulose, hybridized, and washed under stringent conditions. The films were exposed for one day with an intensifying screen at −80°C. A Human Multiple Tissue Northern blot and a Human Immune System Multiple Tissue Northern (Clontech, Palo Alto, CA) were probed with the Exodus cDNA and washed under stringent conditions according to the manufacturer's instructions. The autoradiograph was exposed as above for 1 to 4 days.
Synthetic Exodus production.Exodus was prepared by chemical peptide synthesis using techniques that have been used successfully for the production of other chemokines such as interleukin-8 (IL-8).36 37 Two forms of Exodus (single ala and double ala at nucleotide position 120) were chemically synthesized using optimized stepwise solid-phase methods on an Applied Biosystems 430A Peptide Synthesizer. The synthetic Exodus was purified by reverse-phase HPLC and characterized by standard methods, including electrospray mass spectrometry and nuclear magnetic resonance. Pure synthetic Exodus was reconstituted in phosphate-buffered saline (PBS), and then stored frozen at −70°C in aliquots.
All experiments analyzing the biological activity of Exodus were performed with this pure, quantified, synthetic material. All experimental data reported here are for the single ala form Exodus, which was the original clone isolated. However, all experiments analyzing Exodus activity were repeated with the double ala form of Exodus, and nearly identical results were obtained.
Hematopoietic progenitor assays.Hematopoietic colony formation assays were performed essentially as we previously described.20-23 Volunteer human bone marrow cells were collected from donors after obtaining informed consent. Low density human marrow cells at 5 × 104 mL were plated in 1% methylcellulose in Iscove's modified Dulbecco's medium supplemented with 30% FCS, recombinant human (rHu) erythropoietin (EPO, 1 U/mL), rHu IL-3 (100 U/mL), and rHu SLF (50 ng/mL) for CFU-GM, colony-forming unit granulocyte/erythrocyte/macrophage/megakaryocyte (CFU-GEMM) or burstforming unit-erythrocyte (BFU-E) analysis. Various concentrations of pure synthetic Exodus were compared to purified, quantified members of the beta chemokine family. Cultures were incubated at 5% CO2 and low (5%) oxygen tension for 14 days, and then scored using an inverted microscope. Three plates were scored per data point per experiment.
Chemotaxis assays.Chemotaxis assays were performed as previously described.38 39 Twenty milliliters peripheral blood (PB) was collected from healthy volunteers in 10 mL heparinized tubes. Blood was diluted 1:1 with PBS and then underlaid with 10 mL of Histopaque (Sigma). After centrifugation at 400g for 25 minutes, cells at the interface were collected and washed twice in PBS. Cells were resuspended in Dulbecco's modified Eagle's medium (DMEM) with antibiotics at 106/mL. Sterile bovine serum albumin (BSA) was added to a final concentration of 0.2 mg/mL.
One hundred microliters of the cell suspension was added to each transwell insert (Costar, Cambridge, MA). DMEM with antibiotics and 0.2% BSA with or without pure synthetic Exodus was added to the lower wells in the 24-well plate. All Exodus concentrations were done in triplicate. Transwell inserts were placed into the lower wells, and incubated at 37°C for 90 minutes. At the completion of the incubation period inserts were removed and the top of the filter scraped with a rubber policeman to remove adherent cells. The entire insert was then stained with Wright-Giemsa. Cells adherent to the lower surface of the insert and those that migrated to the lower well were counted under 3 high power fields, and added together to obtain total migrating cells.
RESULTS
Exodus sequence analysis.The complete cDNA of the novel chemokine Exodus is 821 nucleotides in length (Fig 1). There is a consensus polyadenylation site at nucleotide position 786. The 3′ untranslated sequence has a number of AAAU sequences that mediate mRNA stability in many cytokine genes.40 These sequences promote message degradation, and contribute to the short half-life of many cytokine transcripts, including chemokines. There is a short 5′ untranslated region of 43 nucleotides, which contains an in-frame translational stop codon. The sequence 5′ of the ATG that initiates translation fits the Kozak consensus at the first three positions preceding the ATG, but none before that.41 The nucleotide after the ATG does not match the Kozak consensus.
The nucleotide and amino acid sequence of the novel human chemokine Exodus. The predicted signal peptide is underlined. The conserved cysteines that participate in the characteristic disulfide bonds are in bold italics. There are a number of 3′ untranslated AAAU sequences present that often mediate mRNA instability in cytokine genes. There is also a consensus polyadenylation sequence near the 3′ end of the sequence. This sequence has been deposited in GenBank under accession number U64197.
The nucleotide and amino acid sequence of the novel human chemokine Exodus. The predicted signal peptide is underlined. The conserved cysteines that participate in the characteristic disulfide bonds are in bold italics. There are a number of 3′ untranslated AAAU sequences present that often mediate mRNA instability in cytokine genes. There is also a consensus polyadenylation sequence near the 3′ end of the sequence. This sequence has been deposited in GenBank under accession number U64197.
There are 95 amino acids in the conceptual translation of Exodus. This is consistent with the β-chemokine family, where the length of family members ranges from 91 to 99 (Table 1). The first 22 amino acids of Exodus are strongly hydrophobic, and presumably constitute a signal peptide. The four cysteines that participate in the disulfide bonds that define this family are also conserved in Exodus as are five highly conserved residues (Table 1). Exodus is most closely related to MIP-1 α and RANTES at the amino acid level, with 26% to 28% identity, and 75% similarity when conservative changes are taken into account. Exodus is especially similar to RANTES from amino acids 24 to 46 and again from 58 to 75 (numbering from the initiating methionine), where between these positions there are only six nonconservative changes.
Comparison of Exodus With Other β-Chemokines
| Exodus | ESEASNFDCCLGYTDRILHPKFIVGFTRQLANEGCDINAIIFHTKKKLSVCANPKQTWVKYIVRLLSKKVKNM | (100) |
| MIP-1α | ASLAADTPTACCFSYTSRQIPQNFIADY-F-ETSSQCSKPGVIFLTKRSRQVCADPSEEWVQKYVSDLELSA | (28) |
| MIP-1β | APMGSDPPTACCFSYT-REASSNFVVDY-Y-ETSSLCSQPAVVFQTKRSKQVCADPSESWVQEYVYDLELN- | (27) |
| RANTES | SPYSSDT-TPCCFAYIARPLPRAHIKEY-F-YTSGKCSNPAVVFVTRKNRQVCANPEKKWVREYINSLEMS | (26) |
| MCP-1 | QPDAINAPVTCCYNFTNRKISVQRLASY-RRITSSKCPKEAVIFKTIVAKEICADPKQKWVQDSMDHLDKQTQTPKT | (26) |
| MCP-2 | AQPDSVSIPITCCFNVINRKIPIQRLESY-TRITNIQCPKEAVIFKTKRGKEVCADPKERWVRDSMKHLDQIFQNLKP | (22) |
| MCP-3 | QPVGINTSTTCCYRFINKKIPKQRLESY-RRTTSSHCPREAVIFKTKLDKEICADPTQKWVQDFMKHLDKKTQTPKL | (23) |
| MCP-4 | QPDALNVPSTCCFTFSSKKISLQRLKSY-V-ITTSRCPQKAVIFRTKLGKEICADPKEKWVQNYMKHLGRKAHTLKT | (22) |
| I-309 | SKSMQVPFSRCCFSFAEQEIPLRAILCY-R-NTSSICSNEGLIFKLKRGKEACALDTVGWVQRHRKMLRHCPSKRK | (20) |
| Exodus | ESEASNFDCCLGYTDRILHPKFIVGFTRQLANEGCDINAIIFHTKKKLSVCANPKQTWVKYIVRLLSKKVKNM | (100) |
| MIP-1α | ASLAADTPTACCFSYTSRQIPQNFIADY-F-ETSSQCSKPGVIFLTKRSRQVCADPSEEWVQKYVSDLELSA | (28) |
| MIP-1β | APMGSDPPTACCFSYT-REASSNFVVDY-Y-ETSSLCSQPAVVFQTKRSKQVCADPSESWVQEYVYDLELN- | (27) |
| RANTES | SPYSSDT-TPCCFAYIARPLPRAHIKEY-F-YTSGKCSNPAVVFVTRKNRQVCANPEKKWVREYINSLEMS | (26) |
| MCP-1 | QPDAINAPVTCCYNFTNRKISVQRLASY-RRITSSKCPKEAVIFKTIVAKEICADPKQKWVQDSMDHLDKQTQTPKT | (26) |
| MCP-2 | AQPDSVSIPITCCFNVINRKIPIQRLESY-TRITNIQCPKEAVIFKTKRGKEVCADPKERWVRDSMKHLDQIFQNLKP | (22) |
| MCP-3 | QPVGINTSTTCCYRFINKKIPKQRLESY-RRTTSSHCPREAVIFKTKLDKEICADPTQKWVQDFMKHLDKKTQTPKL | (23) |
| MCP-4 | QPDALNVPSTCCFTFSSKKISLQRLKSY-V-ITTSRCPQKAVIFRTKLGKEICADPKEKWVQNYMKHLGRKAHTLKT | (22) |
| I-309 | SKSMQVPFSRCCFSFAEQEIPLRAILCY-R-NTSSICSNEGLIFKLKRGKEACALDTVGWVQRHRKMLRHCPSKRK | (20) |
Alignment of the human members of the beta chemokine family for comparison with the amino acid sequence of Exodus. Exodus is most closely related to MIP-1 α and RANTES. Percent similarity is shown on the right hand side in parentheses. Only the mature product sequence is shown, as signal sequences are poorly conserved among the family.
Although Exodus has many of the conserved amino acid features of the other human β-chemokines (Table 1), there are several unusual characteristics of Exodus that are worth noting. Exodus has a highly basic carboxy terminus, more consistent with the MCP subfamily. In addition, Exodus lacks a conserved tyrosine and a threonine at positions 47 and 51, respectively, which are present in all other human β-chemokines. It is not clear if these two highly conserved amino acids play a role in β-chemokine activity since they are not predicted to be in contact with the receptor.42 When compared to the other beta chemokines, Exodus has one to three additional amino acids in this region between cysteines 2 and 3. These additional residues may compensate for the lack of the conserved amino acids at those sites.
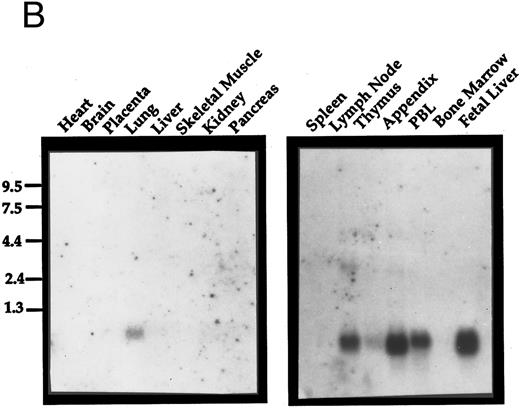
Expression analysis.Exodus has a very restricted pattern of expression. It was not expressed in a number of cell lines tested, including TMR323 neuroblastoma, MDA breast carcinoma, K562 erythroleukemia, Jurkat T-cell leukemia, HL60 promyelocytic leukemia, HL60 cells differentiated to granulocytes with retinoic acid, 3T3 embryonic fibroblasts, or 293 embryonic kidney cells (Fig 2A). When a commercially-prepared Northern blot of a variety of normal human tissues was analyzed for Exodus expression, it was found to be expressed only in the lung (Fig 2B). The size of the transcript was approximately 0.9 kB, consistent with the size of the cDNA reported here.
Northern analysis of the expression of Exodus. (A) Exodus is not expressed in a variety of hematopoietic and nonhematopoietic cell lines in poly A+ Northern analysis. (B) Commercially prepared poly A+ Northern blots of a variety of normal human lymphoid and nonlymphoid tissue. Exodus is preferentially expressed in lymphoid tissue, consistent with the other members of the β-chemokine family. (C) Exodus expression is induced in monocytic and endothelial cells when they are exposed to inflammatory stimuli. PBMC are normal human PBMCs, I-HUVEC are cultured human umbilical vein endothelial cells, and THP-1 are human monocytic leukemia cells. LPS is lipopolysaccharide. The blots in 2A and 2C were rehybridized with actin to control for integrity and loading of RNA. These signals are shown below the Exodus Northerns. The commercially-prepared Northerns were not rehybridized with actin.
Northern analysis of the expression of Exodus. (A) Exodus is not expressed in a variety of hematopoietic and nonhematopoietic cell lines in poly A+ Northern analysis. (B) Commercially prepared poly A+ Northern blots of a variety of normal human lymphoid and nonlymphoid tissue. Exodus is preferentially expressed in lymphoid tissue, consistent with the other members of the β-chemokine family. (C) Exodus expression is induced in monocytic and endothelial cells when they are exposed to inflammatory stimuli. PBMC are normal human PBMCs, I-HUVEC are cultured human umbilical vein endothelial cells, and THP-1 are human monocytic leukemia cells. LPS is lipopolysaccharide. The blots in 2A and 2C were rehybridized with actin to control for integrity and loading of RNA. These signals are shown below the Exodus Northerns. The commercially-prepared Northerns were not rehybridized with actin.
When a commercially prepared Northern blot of a lymphoid tissue was examined for Exodus expression, it was found to be highly expressed in several different lymphoid organs (Fig 2B). Exodus was highly expressed in peripheral lymph nodes, appendix, PBMCs, and fetal liver. It is less highly expressed in the thymus. Interestingly, there was no expression in the spleen or marrow.
Since the expression of many chemokines is induced in mononuclear cells by inflammatory stimuli, the expression of Exodus after LPS, tumor necrosis factor-α (TNF-α) or PMA exposure was analyzed by Northern analysis (Fig 2C). When PBMCs were exposed to LPS for 12 hours Exodus expression was highly induced. This expression declined after 24 hours of exposure to LPS. When umbilical vein endothelial cells were exposed to TNF-α for just 3 hours, Exodus expression was again highly induced. Significantly, Exodus expression stayed high as long as there was the inflammatory stimuli present. When the monocytic leukemia cell line THP-1 was treated with PMA the expression of Exodus was also induced, reaching its peak at 48 hours after exposure, and declining slightly thereafter.
Inhibition of hematopoietic progenitors.We previously reported that beta chemokines negatively regulated the proliferation of immature subsets of hematopoietic progenitors.20-23 To test whether Exodus also inhibited proliferation of hematopoietic progenitors we performed colony formation assays in the presence or absence of pure synthetic Exodus (Table 2). Culture medium alone or various chemokines known not to have inhibitory activity served as negative controls. Chemokines known to have inhibitory activity served as positive controls.
Inhibition of Human Hematopoietic Progenitors by Exodus
| . | CFU-GM . | BFU-E . | CFU-GEMM . |
|---|---|---|---|
| Medium | 85 ± 3 | 97 ± 4 | 39 ± 3 |
| rHu Exodus (200*) | 43 ± 11 | 43 ± 2 | 19 ± 3 |
| rHu Exodus (100) | 39 ± 3 | 41 ± 2 | 20 ± 2 |
| rHu Exodus (50) | 42 ± 10 | 42 ± 3 | 17 ± 2 |
| rHu Exodus (25) | 41 ± 2 | 50 ± 2 | 20 ± 4 |
| rHu Exodus (12.5) | 51 ± 3 | 70 ± 10 | 27 ± 1 |
| rHu Exodus (6.25) | 64 ± 8 | 87 ± 3 | 32 ± 2 |
| rHu MIP-1 α (100) | 41 ± 2 | 44 ± 3 | 19 ± 2 |
| rHu IL-8 (100) | 42 ± 2 | 44 ± 2 | 19 ± 2 |
| rHu PF-4 (100) | 42 ± 4 | 44 ± 5 | 19 ± 1 |
| rHu RANTES (100) | 81 ± 7 | 99 ± 4 | 37 ± 1 |
| rHu NAP-2 (100) | 83 ± 2 | 93 ± 3 | 39 ± 4 |
| . | CFU-GM . | BFU-E . | CFU-GEMM . |
|---|---|---|---|
| Medium | 85 ± 3 | 97 ± 4 | 39 ± 3 |
| rHu Exodus (200*) | 43 ± 11 | 43 ± 2 | 19 ± 3 |
| rHu Exodus (100) | 39 ± 3 | 41 ± 2 | 20 ± 2 |
| rHu Exodus (50) | 42 ± 10 | 42 ± 3 | 17 ± 2 |
| rHu Exodus (25) | 41 ± 2 | 50 ± 2 | 20 ± 4 |
| rHu Exodus (12.5) | 51 ± 3 | 70 ± 10 | 27 ± 1 |
| rHu Exodus (6.25) | 64 ± 8 | 87 ± 3 | 32 ± 2 |
| rHu MIP-1 α (100) | 41 ± 2 | 44 ± 3 | 19 ± 2 |
| rHu IL-8 (100) | 42 ± 2 | 44 ± 2 | 19 ± 2 |
| rHu PF-4 (100) | 42 ± 4 | 44 ± 5 | 19 ± 1 |
| rHu RANTES (100) | 81 ± 7 | 99 ± 4 | 37 ± 1 |
| rHu NAP-2 (100) | 83 ± 2 | 93 ± 3 | 39 ± 4 |
Colony formation assays of the effect of recombinant human Exodus on the proliferation of hematopoietic progenitors. There was a dose-dependent inhibition of hematopoietic progenitor proliferation by Exodus that was comparable to the inhibitory activities of other β-chemokines known to inhibit the proliferation of hematopoietic progenitors. Experiments were performed twice in triplicate. The values are the mean plus or minus the standard error. There is a statistically significant (Student's t-test) reduction in colony formation at concentrations of Exodus down to 25 ng/mL (P < .005).
Concentrations in ng/mL.
Exodus inhibited hematopoietic progenitor colony formation in a dose-dependent manner. There was no inhibitory activity of hematopoietic progenitors with medium alone or with RANTES or NAP-2. At 100 ng/mL of recombinant human MIP-1 α or IL-8, a dose at which the biological effect plateaus, there was a statistically significant reduction of CFU-GM (48% of medium control for MIP-1 α and 49% for IL-8), BFU-E (45% of control for both), and CFU-GEMM (49% of control for both). Each of these decreases were statistically significant using Student's t-test.
Using ascending concentrations of pure synthetic Exodus a dose response in hematopoietic progenitor inhibition was seen. Exodus inhibitory activity plateaus at 50 ng/mL, where there was also a statistically significant decrease in CFU-GM (49% of control), BFU-E (43% of control), and CFU-GEMM (43% of control).
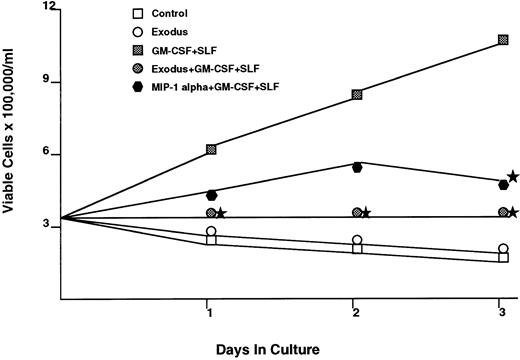
The effect of Exodus on the proliferation of cytokine dependent myeloid cell lines was also tested (Fig 3). The human myeloid cell line MO7E requires GM-CSF and SLF for maximal proliferation. When synthetic pure human Exodus is added to the log phase MO7E cells, in the presence of GM-CSF and SLF, proliferation over the next 72 hours is reduced to 10.4% of control (GM-CSF plus SLF plus equivalent amount of pECE/Cos supernatant). It should be noted that this was not a cytotoxic effect, as the Exodus-treated cells had greater than 95% viability at every time point, identical to that of the control cells. This reduction is statistically significant using Student's t-test at P < .05.
The effect of pure synthetic Exodus on the cytokine-dependent human myeloid progenitor cell line MO7E.43 This cell line requires GM-CSF and SLF for maximal proliferation. The addition of Exodus almost completely inhibited proliferation. However, viability as assessed by trypan blue exclusion did not change with the addition of Exodus. Each data point is the mean of six separate experiments performed in triplicate. Stars denote statistical significance at P < .05 using a paired t-test between the Exodus-treated MO7e cells in GM-CSF/SLF and MO7e cells only in GM-CSF/SLF.
The effect of pure synthetic Exodus on the cytokine-dependent human myeloid progenitor cell line MO7E.43 This cell line requires GM-CSF and SLF for maximal proliferation. The addition of Exodus almost completely inhibited proliferation. However, viability as assessed by trypan blue exclusion did not change with the addition of Exodus. Each data point is the mean of six separate experiments performed in triplicate. Stars denote statistical significance at P < .05 using a paired t-test between the Exodus-treated MO7e cells in GM-CSF/SLF and MO7e cells only in GM-CSF/SLF.
Chemotaxis.Pure synthetic Exodus was analyzed for the ability to stimulate chemotaxis of normal human PBMCs using transwell migration assays (Fig 4). MIP-1 α served as the positive control and medium alone as the negative control. Medium alone stimulated an average of 7 cells per high powered field to migrate acrossed the transwell insert. Maximal concentrations of MIP-1 α stimulated an 18-fold increase in mononuclear cell chemotaxis, to an average of 128 cells per high powered field. Escalating concentrations of pure synthetic Exodus all stimulated chemotaxis of the mononuclear cells across the transwell inserts. Five hundred nanograms per milliliter of Exodus produced a 25-fold rise in chemotaxis, to 178 cells. The migrating cell number stimulated by all concentrations of Exodus was statistically significantly greater than the negative control in unpaired Student's t-test at P < .05. In addition, Exodus appears to be more effective at stimulating chemotaxis than MIP-1 α at its maximal effective concentration.
The effect of pure synthetic Exodus on chemotactic activity of normal human PBMCs. Chemotaxis was measured by transwell migration. The highest concentrations of Exodus stimulated chemotaxis more efficiently than maximally effective concentrations of MIP-1 α. The values represent the average of two experiments performed in nine transwells each time plus or minus the standard error. The stars represent statistically significant differences from the control at P < .05 using the unpaired Student's t-test.
The effect of pure synthetic Exodus on chemotactic activity of normal human PBMCs. Chemotaxis was measured by transwell migration. The highest concentrations of Exodus stimulated chemotaxis more efficiently than maximally effective concentrations of MIP-1 α. The values represent the average of two experiments performed in nine transwells each time plus or minus the standard error. The stars represent statistically significant differences from the control at P < .05 using the unpaired Student's t-test.
DISCUSSION
We have isolated and characterized a novel member of the β-chemokine family, termed Exodus. Although Exodus shares many of the conserved sequence features of this family, it has some notable exceptions. Exodus does not have a tyrosine at position 47 or a threonine at position 50. These amino acids are seen in all other members of this family. These residues may not interact with chemokine receptors based on the crystallographic structures of MIP-1 α, MIP-1 β, and RANTES.42 However, they may still be important in the overall function of this family, perhaps by maintaining appropriate tertiary structure, or mediating dimerization. Exodus has similarity to the other beta chemokine family members in the positioning of the conserved cysteines that participate in the disulfide bonds that define this family.
Exodus is more divergent from the other β-chemokines based on amino acid sequence than they are from each other. It is possible that Exodus may have some unique functional properties that distinguish it from the rest of this group, based on these distinct structural differences. Because Exodus sequence is more divergent from the other β-chemokines, yet it shares many of the same biological activities, it is possible that Exodus is the prototype of a divergent branch of the β-chemokine family, like the MCP.19 We recently have obtained preliminary evidence that this may be the case, by isolating two other novel, uncharacterized β-chemokine cDNAs that are more related to Exodus than any other β-chemokine.
Exodus is expressed in a pattern typical of many chemokines.1 Northern analysis showed that Exodus was expressed mainly in lymphoid tissue, especially in lymph nodes, the appendix, and PB. The in vitro experiments indicated that Exodus was poorly expressed unless inflammatory stimuli was present. This upregulation on mononuclear cell activation is typical of many chemokine family members.1 Once an immunologic stimulus was present Exodus was rapidly upregulated. The nature of the stimulus itself also seemed unrestricted, with LPS, TNF-α, and PMA all upregulating Exodus. However, only TNF-α produced prolonged Exodus expression, implying that there may be multiple pathways for the regulation of Exodus expression. In vivo Exodus may be regulated by normal immunologic stimuli such as cytokine activation.
Exodus was poorly expressed in marrow as opposed to PB. This may be secondary to the fact that there the marrow is mainly composed of immature myeloid and erythroid precursors, while there are far more mature mononuclear cells in the PB. The Northern analysis with the cell lines shows that Exodus is expressed preferentially in activated cells of monocytic origin. Exodus production appears to be a function of a mature lymphophagocytic cell as opposed to immature myeloid cells.
Although Exodus was isolated from pancreatic islet cell cDNA it was poorly expressed in pancreatic tissue. Perhaps there was mononuclear cell contamination of the islets, or isolation of the Exodus cDNA was fortuitous, in which case a very low abundance transcript was happened upon.
We and others had previously found that beta chemokines such as MIP-1 alpha inhibited hematopoietic proliferation at a very primitive level.19-22 Previous data had suggested that β-chemokines had a negative regulatory effect on stem/progenitor cells. This was not a cytotoxic effect, but rather a cell cycle arrest.22 As such, pre-exposure with β-chemokines such as MIP-1 α could be chemoprotective. Cell cycle-specific chemotherapeutic agents were not as cytotoxic to murine marrow when that marrow had been pretreated with a β-chemokine.
In this study we found that recombinant Exodus also inhibited proliferation of hematopoietic progenitors. Indeed, this inhibition was perhaps slightly more effective than MIP-1 α. This raises the possibility that Exodus may have clinical utility in some hematologic disorders.23
In addition, it may have a relevant normal physiological role in negatively regulating marrow proliferation as well, when marrow proliferation is no longer needed, as when recovery from infection or trauma occurs. Slowing marrow progenitor proliferation may turn out to be as important a control of normal hematopoiesis as apoptosis. Infection could be viewed as producing its own negative regulatory feedback loop on hematopoiesis by stimulating production of chemokines such as MIP-1 α and Exodus. Certainly, based on the expression analysis of Exodus, inflammatory states would increase the production of Exodus by mononuclear cells.
Exodus also shares the biological activity classically associated with beta chemokines. It stimulates the chemotaxis of PBMCs. Thus, Exodus activities are similar to MIP-1 α, the β-chemokine with which it shares homology. Exodus may perhaps be even more effective than MIP-1 α at promoting chemotaxis, based on the transwell migration assays used here.
In this report the isolation and initial characterization of a novel β-chemokine, termed Exodus, was described. Although the structure is more divergent than other β-chemokines, its activity and patterns of expression are similar to other members of the β-chemokine family. Based on the activities characterized here it may be have clinical utility in (1) chemoprotection of marrow, or (2) against myeloproliferative diseases. Because Exodus shares some of the activities of MIP-1 α it will be especially intriguing to investigate its activity against HIV.
Address reprint requests to Robert Hromas, MD, Department of Hematology/Oncology at the Walther Oncology Center, Indiana University Medical Center, IB442, 975 W Walnut St, Indianapolis, IN 46202.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal