Abstract
Platelet glycoprotein (GP) V is a major surface protein cleaved during thrombin-induced platelet activation. GPV associates noncovalently with the GPIb-IX complex to form GPIb-V–IX, a receptor for von Willebrand factor and thrombin. We describe the cloning of the genes coding for rat and mouse GPV and compare them with the human gene. The two rodent genes have a similar structure and resemble the human GPV gene with a coding sequence (≈1,700 nucleotides) entirely contained in one exon and a single intron (≈900 nucleotides) in the 5′ untranslated region. Both genes have megakaryocyte-type promoters with conserved tandem Ets and GATA recognition motifs and lack a TATA box. The mature rat and mouse proteins comprise 551 amino acids, have 70% sequence identity, and contain an additional 8–amino acid intracellular segment as compared with the human protein. As in human GPV, there is an NH2 -terminal leucine-rich region of 15 repeats and a thrombin cleavage recognition sequence. Whereas the rat and human thrombin cleavage sites are similar, the mouse cleavage site resembles that of the human thrombin receptor. Functionality of these sites was demonstrated by thrombin cleavage of synthetic peptides and analysis by high-performance liquid chromatography (HPLC) or mass spectrometry. Cleavage of native rat GPV was confirmed by means of a polyclonal antibody directed against the new NH2 -terminal peptide exposed after thrombin cleavage. This antibody specifically recognized thrombin-activated rat platelets by fluorescence-activated cell sorting (FACS) analysis. In addition, we raised monoclonal antibodies specific for rat GPV (88 kD), which recognized the NH2 -terminal soluble fragment (70 kD) liberated after thrombin cleavage. Knowledge of these rodent GPV genes and availability of species-specific peptides and antibodies will be essential to further studies aiming to define the exact in vivo function of platelet GPV using animal models of thrombosis and gene inactivation experiments.
THE ADHESIVE properties of platelets are mediated by two main surface receptors that belong to the integrin and leucine-rich families of adhesive receptors.1,2 The latter is represented in platelets by a four-subunit receptor, the glycoprotein (GP) Ib-V–IX complex. This receptor is formed by the disulfide-bonding of GPIbα (145 kD) and GPIbβ (24 kD) to form GPIb, which associates noncovalently with GPIX (20 kD) in 1:1 stoichiometry and with GPV (82 kD) in 2:1 stoichiometry.3,4 GPIb-V–IX is a receptor for von Willebrand factor, and is involved in platelet adhesion to the subendothelium5 and in platelet activation under high shear conditions.6 Over the years, detailed studies of GPIbα have led to the identification of binding sites for von Willebrand factor and thrombin.7 8 Conversely, less is known about the exact functions of GPIbβ, GPIX, and GPV.
Cloning and sequencing of the cDNAs and genes coding for all four subunits forming GPIb-V–IX has been completed.9-16 We have characterized the gene for human GPV, which comprises a short intron and a coding sequence contained entirely in the second exon.16 Mature GPV has 544 amino acids, a single transmembrane domain at the COOH terminus, and a 16–amino acid cytoplasmic tail. Its extracellular domain contains 15 tandem Leu-rich repeats of 24 amino acids and a cleavage site for thrombin near the transmembrane domain. The GPV promoter contains several GATA and ets cis-acting elements common to all megakaryocytic specific genes described to date.10 15-19
A certain number of questions regarding the biosynthesis of GPV and its functional role in hemostasis remain unanswered. Transfection experiments have shown that efficient expression of GPIbα requires the presence of the GPIbβ and GPIX subunits and is further increased by the presence of GPV.20-22 These experiments also pointed to a direct association between GPIbα and GPV. A role for GPV in thrombin-induced platelet aggregation was initially suggested on the basis of its specific cleavage by thrombin.23 This proposition has been challenged based on the lack of effect of GPV blocking antibodies24 and GPV cleavage by other proteases25 on thrombin-induced aggregation. Subsequently, a G-protein coupled receptor involved in thrombin activation has been characterized.26 27 GPV could still be important in modulating the activation response by binding to thrombin, or could play a role in fibrinolysis or angiogenesis.
One possible strategy to define the exact in vivo function of GPV would be to test its effect in mouse and rat models of thrombosis and thrombolysis or to inactivate its gene in mice. However, this requires prior knowledge of the sequence in these species and the development of species specific reagents such as recombinant proteins and antibodies. In this report, we present the cloning and sequencing of the rat and mouse GPV genes. Promoter regions and deduced protein sequences were compared with those of the human gene and specific rat peptides and antibodies were developed. The promoters contain possible binding sites for megakaryocyte transcription factors, while the proteins are well conserved especially in the Leu-rich region and contain a cleavage site for thrombin. Functionality of the cleavage site was tested using synthetic peptides for all three species and the native rat protein with species-specific antibodies.
Characterization of the rat and mouse platelet GPV genes. (A) Cloning strategy and structure of the rat and mouse GPV genes and comparison to the human gene. The rat GPV gene was obtained by screening of a λ Dash genomic library (λVR1 clone) with a human genomic DNA probe, PCR amplification of rat genomic DNA, and rescreening of the genomic library (λVR2 clone) with the PCR fragment. The reported sequence corresponds to the λVR2 clone and contains the entire coding sequence and promoter region. The mouse GPV gene (λVM1 clone) was obtained by screening a mouse genomic library in the λ Dash vector with the rat λVR1 clone. Each clone was sequenced at least once on both strands. The human GPV gene structure16 is given for comparison. The three genes have a single intron dividing the genes in two exons. The clones and intron sizes are indicated in nucleotides (nt). Stippled, closed, and open boxes represent the coding, 5′ and 3′ untranslated, and promoter regions, respectively. The ATG start and TGA stop codons are marked with (▴). The (▵) indicates the position of a putative transcription start site. Sites for EcoRI, Xho I, HindIII, Pst I, and BamHI restriction enzymes are indicated. The EcoRI site in parentheses is a cloning site not present in the gene. (B) Comparison of human (H), rat (R), and mouse (M) GPV promoter sequences. Approximatly 500 nt of the 5′ flanking sequences upstream of the intron are shown. Nucleotide numbering on the left corresponds to Lanza et al16 for human GPV and to the clones λRV2 and λMV1 for rat and mouse GPV, respectively. A putative initiation start site is boxed. Negative nucleotide numbering on the basis of the proposed initiation site is indicated on the right. Identity with the human sequence is indicated by a (:), while hyphens denote gaps inserted to optimize the alignment. Putative binding sites for TATA, GATA, Ets, PuF, CACC, NFE-2, and the platelet specific PMS-E transcription factors are indicated by bold-underlined characters. (C) Transcriptional start sites in megakaryocyte and platelet expressed genes. The sequences flanking the +1 position for mouse terminal deoxynucleotidyl transferase (TdT) and human αIIb and α2 integrins, GPIX, and PF4 genes are compared with 5′ flanking sequences of H, R, and M GPV genes. Sites of transcription initiation for promoters lacking TATA and CAAT boxes have a consensus sequence with CA at −1, +1 (boxed) and pyrimidine-rich segments on either side (underlined).
Characterization of the rat and mouse platelet GPV genes. (A) Cloning strategy and structure of the rat and mouse GPV genes and comparison to the human gene. The rat GPV gene was obtained by screening of a λ Dash genomic library (λVR1 clone) with a human genomic DNA probe, PCR amplification of rat genomic DNA, and rescreening of the genomic library (λVR2 clone) with the PCR fragment. The reported sequence corresponds to the λVR2 clone and contains the entire coding sequence and promoter region. The mouse GPV gene (λVM1 clone) was obtained by screening a mouse genomic library in the λ Dash vector with the rat λVR1 clone. Each clone was sequenced at least once on both strands. The human GPV gene structure16 is given for comparison. The three genes have a single intron dividing the genes in two exons. The clones and intron sizes are indicated in nucleotides (nt). Stippled, closed, and open boxes represent the coding, 5′ and 3′ untranslated, and promoter regions, respectively. The ATG start and TGA stop codons are marked with (▴). The (▵) indicates the position of a putative transcription start site. Sites for EcoRI, Xho I, HindIII, Pst I, and BamHI restriction enzymes are indicated. The EcoRI site in parentheses is a cloning site not present in the gene. (B) Comparison of human (H), rat (R), and mouse (M) GPV promoter sequences. Approximatly 500 nt of the 5′ flanking sequences upstream of the intron are shown. Nucleotide numbering on the left corresponds to Lanza et al16 for human GPV and to the clones λRV2 and λMV1 for rat and mouse GPV, respectively. A putative initiation start site is boxed. Negative nucleotide numbering on the basis of the proposed initiation site is indicated on the right. Identity with the human sequence is indicated by a (:), while hyphens denote gaps inserted to optimize the alignment. Putative binding sites for TATA, GATA, Ets, PuF, CACC, NFE-2, and the platelet specific PMS-E transcription factors are indicated by bold-underlined characters. (C) Transcriptional start sites in megakaryocyte and platelet expressed genes. The sequences flanking the +1 position for mouse terminal deoxynucleotidyl transferase (TdT) and human αIIb and α2 integrins, GPIX, and PF4 genes are compared with 5′ flanking sequences of H, R, and M GPV genes. Sites of transcription initiation for promoters lacking TATA and CAAT boxes have a consensus sequence with CA at −1, +1 (boxed) and pyrimidine-rich segments on either side (underlined).
MATERIALS AND METHODS
Materials.Nitrocellulose filters were obtained from Millipore, St Quentin en Yvelines, France and [32P] dCTP (deoxycytidine 5′-triphosphate), [35S] dATP (deoxyadenosine 5′-triphosphate), and the Sequenase II sequencing kit from Amersham, Les Ulis, France. Restriction enzymes were purchased from New England Biolabs, Ozyme, Montigny Le Bretonneux, France. Oligonucleotide primers were synthesized using an Oligo 1000 synthesizer (Beckman, Gagny, France). Peptides were synthesized and coupled to keyhole limpet hemocyanin (KLH) by Neosystem SA, Strasbourg, France. Purified human α-thrombin was obtained from the Etablissement de Transfusion Sanguine de Strasbourg, Strasbourg, France. Recombinant hirudin was a gift of Dr A. Pavirani, Transgène SA, Strasbourg, France. Aprotinin, aPMSF ([4-amidinophenyl]-methane sulfonylfluoride), leupeptin, M13 vector, Taq polymerase, and Nutridoma-CS medium were purchased from Boehringer Mannheim, Germany. PGI2 was obtained from Sigma Chemicals, St Louis, MO, heparin from Hoffmann La Roche, Basel, Switzerland, and Pefablock from Interchim, Montluçon, France. Fluorescein isothiocyanate (FITC)-conjugated antirabbit F(ab)′2 and FITC-conjugated goat antimouse F(ab)′2 antibodies were purchased from Immunotech, Luminy, France. Horseradish peroxidase (HRP)-coupled antirabbit immunoglobulins (Igs) were obtained from Bio-Rad Laboratories, Hercules, CA and polyethylene glycol (PEG), Dulbecco's modified Eagle's medium (DMEM), NCTC-135, and hypoxanthine, aminopterin, thymidine (HAT) from Life Technologies, Eragny, France.
Animals.New Zealand White rabbits and 6- to 8-week-old female Balb/c mice were purchased from the Centre de Production Animale, Olivet, France and Wistar rats from IFA-CREDO, Arbresles, France.
Cells.The murine myeloma cell line P3x653.Ag8 was a generous gift from Dr J.E.K. Hildreth, The Johns Hopkins University School of Medicine, Baltimore, MD. Hybridoma and the myeloma cell lines were maintained in HY media (DMEM supplemented with 10% (vol/vol) fetal bovine serum (FBS), 10% (vol/vol) NCTC-135, 10 mmol/L HEPES, 0.2 U/mL insulin, 0.45 mmol/L pyruvate, and 1 mmol/L oxaloacetate). Cells were grown at 37°C in a humidified 5% CO2 atmosphere.
Isolation and characterization of the rat and mouse GPV genes.A human GPV gene fragment corresponding to nucleotides 2,815-3,54916 was labeled with [32P] by random priming and used to screen a rat genomic library in the λ Dash vector (Stratagene, La Jolla, CA). Nitrocellulose filters were hybridized overnight at 42°C in 30% (vol/vol) formamide, 5× SSC, 0.1% (wt/vol) sodium dodecyl sulfate (SDS), 5× Denhardt's medium, and 0.1 mg/mL salmon sperm DNA. The filters were washed twice at 50°C for 30 minutes in 2× SSC, washed twice at 50°C for 30 minutes in 0.05% (wt/vol) SDS and 0.5× SSC, and exposed for autoradiography. One positive clone was detected and subjected to two additional rounds of screening to isolate a 7.9-kb clone. After restriction enzyme digestion and Southern blot analysis, a single EcoRI 2.2-kb fragment was subcloned in the pBluescript vector for further restriction enzyme analysis and finally subcloned in M13 for nucleotide sequencing. To extend the sequence in the 5′ direction, polymerase chain reaction (PCR) amplification was performed on 100 ng of rat leukocyte genomic DNA. PCR conditions were: 0.2 μmol/L of each nucleotide pair, 1 U of Taq polymerase, denaturation at 94°C for 4 minutes and amplification for 30 cycles with extension at 72°C for 1.5 minutes, denaturation at 94°C for 1 minute, and primer annealing at 55°C for 1 minute. A mouse genomic library in the λ Dash vector (Stratagene) was screened with a 2.2-kb rat GPV gene fragment (nt 1,341-3,573). Screening was identical except for the following modifications: 50% (vol/vol) formamide for hybridization and one final wash at 55°C for 30 minutes in 0.5 × SSC, 0.05% (wt/vol) SDS. One positive 18.3-kb clone was obtained and mapped by restriction enzyme analysis. A 3.5-kb EcoRI fragment containing the coding sequence was subcloned in the M13 vector for nucleotide sequencing, which was performed on both strands using the Sequenase II kit. The sequences were assembled and analyzed with the PC Gene software (Intelligenetics, Mountain View, CA) using TRANSL for coding sequence determination, CLUSTAL for multisequence alignment, and FASTA for homology calculation. The rat and mouse GPV gene sequences reported here have been submitted to the Genbank/EMBL Databank with accession numbers Z69594 and Z69595.
Testing for thrombin cleavage of GPV synthetic peptides by high performance liquid chromatography (HPLC) and mass spectrometry.Synthetic peptides corresponding to human (HV1: PGPRGPPPRPAADSY) and mouse (MV1: PDPRSLPLDPPTENY) GPV473-486 (100 μmol/L) were incubated with 40 U/mL human α-thrombin at 37°C for 1 to 12 hours in 50 mmol/L Tris buffer, pH 8, while rat (RV1: PSSRGLPPDPPTENY) GPV473-486 (100 μmol/L) was incubated with 100 U/mL α-thrombin. In HPLC experiments, the samples were then treated at 4°C for 1 hour with 10% (vol/vol) trichoroacetic acid (TCA) to precipitate thrombin and centrifuged at 12,000g for 3.5 minutes at room temperature. Cleavage was assessed by analysis on an HPLC LKB1250 apparatus (LKB, Broma, Finland) using a Nucleosil C18 N125 column (Shandon, Cergy Pontoise, France) with a 5% to 65% acetonitrile gradient in water and elution was monitored at 210 nm wavelength. Cleavage was controlled using synthetic uncleaved 473-486 peptides and short 477-486 peptides as standards.
Matrix assisted laser desorption ionization mass spectrometry (MALDI-MS) experiments were performed on a Bruker (Bremen, Germany) BIFLEXTM time-of-flight mass spectrometer.28 Samples were prepared by deposing first a uniform layer of fine granular matrix crystals using the fast evaporation method of Vorm and Mann.29 A second layer containing the peptide sample was added by successive deposition of 0.8 μL of 2% aqueous trifluoroacetic acid (TFA), 0.5 μL of peptide, and a 0.2-μL aliquot of the matrix (α-cyano-3-hydroxy-trans-cinnamic acid dissolved in acetonitrile/water (2/1) at a concentration of ≈20 mg/mL), followed by evaporation of the solvent at room temperature. Samples were desorbed/ionized using a pulsed nitrogen laser beam (l = 337 nm) at a repetition rate of 1.5 Hz, and the spectra obtained usually corresponded to averages of 50 to 100 laser shots. All studies were performed in the positive and negative modes at an ion acceleration potential of 30 kV. MALDI mass spectra were externally calibrated using the [M+H]+ ions of angiotensin II (1047.20 Da), ACTH 18-39 (2466.73 Da) and bovine insulin (5734.56 Da), and/or using matrix peaks.
Shared Identity of DNA and Predicted Amino Acid Sequences for Human, Rat, and Mouse GPV
| . | DNA (%) . | Amino Acids (%) . | |||||
|---|---|---|---|---|---|---|---|
| . | Coding . | Intron . | Promoter . | Coding . | LR . | LR → TM . | TM + IC . |
| Human/rat | 78 | 61 | 71 | 71 | 77 | 52 | 67 |
| Human/mouse | 79 | 57 | 73 | 70 | 76 | 49 | 69 |
| Rat/mouse | 91 | 81 | 89 | 86 | 88 | 78 | 84 |
| . | DNA (%) . | Amino Acids (%) . | |||||
|---|---|---|---|---|---|---|---|
| . | Coding . | Intron . | Promoter . | Coding . | LR . | LR → TM . | TM + IC . |
| Human/rat | 78 | 61 | 71 | 71 | 77 | 52 | 67 |
| Human/mouse | 79 | 57 | 73 | 70 | 76 | 49 | 69 |
| Rat/mouse | 91 | 81 | 89 | 86 | 88 | 78 | 84 |
Pairwise % identity of DNA and amino acid sequences was determined using the FASTA program from the PC GENE software. Genomic DNA regions comprising the entire coding sequence, the intron, and the promoter region were aligned. The 5′ flanking promoter region, which was analyzed contained the first 200 nucleotides preceding the putative transcription initiation site (Fig 1B). Amino acids from the entire coding region or from portions corresponding to the Leu-rich domain (LR), the region connecting the Leu-rich domain to the membrane (LR → TM), and the region comprising the transmembrane and intracellular domains (TM + IC) were aligned.
Preparation of rat platelet.Wistar rats were anesthesized and blood was collected from the aorta into a syringe containing 1/6 vol of acid-citrate-dextrose (ACD) anticoagulant supplemented with 100 U/mL hirudin, 1 mmol/L EDTA, and 0.1 mmol/L aPMSF. Platelet-rich plasma was obtained by centrifugation at 1,570g for 2.5 minutes. Platelets were pelleted, washed three times in 10 mmol/L HEPES, pH 7.6, 150 mmol/L NaCl, 1 mmol/L EDTA, 0.01 mmol/L aPMSF, 7.5 nmol/L PGI2,30 and finally suspended in the same buffer without PGI2 for use in flow cytometry or lysed with SDS for polyacrylamide gel electrophoresis (PAGE) and Western blot analysis.
Purification of rat GPV.Rat GPV was purified from platelets according to a procedure used to purify human GPV.31 Washed rat platelets (2 × 1012) were incubated at 37°C for 16 hours in 10 mmol/L HEPES, pH 7.6, 300 mmol/L NaCl, 1 mmol/L EDTA, and 0.01 mmol/L aPMSF. The extract was brought to 40% saturation with ammonium sulfate and centrifuged at 11,000g for 20 minutes. This supernatant was then brought to 60% saturation with ammonium sulfate and centrifuged at 11,000g for 20 minutes. The resulting pellet was dissolved in 50 mmol/L potassium phosphate, pH 6.8, 1 mmol/L EDTA (Buffer A), and frozen at −80°C until further purification. Pooled platelet extracts from 1,000 rats were dialyzed against Buffer A and concentrated. Samples were applied to a Superose 12 HR 10/30 gel filtration column (Pharmacia, Orsay, France) and eluted with Buffer A at 0.5 mL/minute. Glycoprotein-containing fractions were then applied to a Mono Q-HR column (Pharmacia) in Buffer A. The flow-through was applied to a wheat germ agglutinin-Sepharose column (Pharmacia) in Buffer A and proteins were eluted with 2.5% (wt/vol) N-acetyl glucosamine in Buffer A. The final yield from 2 × 1012 platelets was 200 μg of GPV, a value comparable to that reported for human platelet GPV.31
Production of monoclonal antibodies (MoAbs) and antipeptide polyclonal Abs against rat GPV.Female Balb/c mice (6 to 8 weeks old) were injected intraperitoneally (IP) with 109 washed rat platelets three times at 3-week intervals. One week after the third injection, the mice were injected IP with 108 washed rat platelets and intravenously (IV) with 108 washed rat platelets. Four days later, the spleen of one mouse was removed and fused with the myeloma cell line P3x653.Ag8 as previously described32 using 50% (wt/vol) PEG with 5% (vol/vol) dimethyl sulfoxide (DMSO). Fusion was screened by flow cytometry as described below, and hybridomas secreting antibodies of interest were subcloned twice by limiting dilution. Antibodies against synthetic rat GPV477-490 (RV4: GLPPDPTENALKAY) peptides were produced in rabbits using the following protocol: 100 μg KLH-peptide in complete Freund adjuvant was injected subcutaneously (SC), followed by two injections SC in incomplete Freund adjuvant. After 3 days, three booster injections of 100 μg KLH-peptide were given 3 to 4 days apart in the ear vein. Blood samples were taken 4 to 7 days after the last injection and weekly thereafter.
Flow cytometric analysis of rat platelet GPV.Analyses were performed on washed rat platelets (2 × 105 platelets in 200 μL) with or without stimulation in the presence of 5 U/mL human α-thrombin at 37°C for 5 minutes followed by neutralization with 10 U/mL hirudin. The cells were incubated with purified antibodies (2 μg) or hybridoma culture supernatants (100 μL) at 22°C for 30 minutes, washed in 10 mmol/L HEPES, pH 7.6, 150 mmol/L NaCl, and 1 mmol/L EDTA and incubated in the dark at 22°C for 30 minutes with 1 μg FITC-conjugated goat antirabbit F(ab)′2 or 1 μg FITC-conjugated goat antimouse F(ab)′2 antibodies. Platelets were finally resuspended in the same buffer and analyzed on a FACSort fluorescence cytometer (Becton Dickinson, San Jose, CA) using Cell Quest software. The light scattering and fluorescence intensity from 10,000 platelets was collected using a logarithmic gain.
Immunoprecipitation of human and rat platelet surface proteins.Human and rat platelets were labeled with sulfo-N-hydroxysuccinimide-biotin (NHSS-biotin, Pierce Chemicals, Rockford, IL) as previously described.33 Briefly, 1 × 109 washed platelets were treated for 30 minutes at 0°C in phosphate-buffered saline (PBS) with 10 mmol/L NHSS-biotin. The biotin-labeled platelets were washed three times with PBS containing 5 mmol/L EDTA and lysed in Tris-EDTA-NaCl (TEN) buffer (50 mmol/L Tris, pH 7.5, 5 mmol/L EDTA, and 150 mmol/L NaCl) containing 1% (vol/vol) Triton X-100 and a protease inhibitor cocktail (0.5 mmol/L PMSF, 1 μg/mL aprotinin, 1 μg/mL leupeptin, 0.5 μg/mL chymostatin, and 0.5 μg/mL antipain) for 2 hours at 0°C. This lysate was precleared with 5 μL normal mouse serum, 10 μg rabbit antimouse IgG (rabbit antimouse, RAM; Jackson Immunoresearch, West Grove, PA) and 50 μL formalin-fixed Staphylococcus aureus Cowen strain I (SaC; Pansorbin, Calbiochem, La Jolla, CA). Precleared lysate was incubated successively with MoAbs for 1 hour at 0°C and with 10 μg RAM IgG for 1 hour at 0°C. Immune complexes were precipitated by addition of SaC to a final concentration of 2%, the mixture was incubated for 30 minutes at 0°C, and the SaC pellet was washed with TEN lysis buffer. Pellets were taken up in SDS-PAGE sample buffer and incubated at 100°C for 3 minutes. The eluted proteins were separated by SDS-PAGE and transferred to nylon membranes. The membranes were blocked with 5% (wt/vol) dried milk in PBS for 1 hour at 20°C, washed three times with PBS-T (PBS containing 0.05% (vol/vol) Tween-20) and incubated with a 1:10,000 dilution of streptavidin-HRP (Pierce) in PBS-T containing 0.1% (wt/vol) bovine serum albumin (BSA) for 1 hour at 20°C. Finally, the membranes were washed three times in PBS-T and biotinylated proteins were detected using a chemiluminescent substrate (ECL, Amersham).
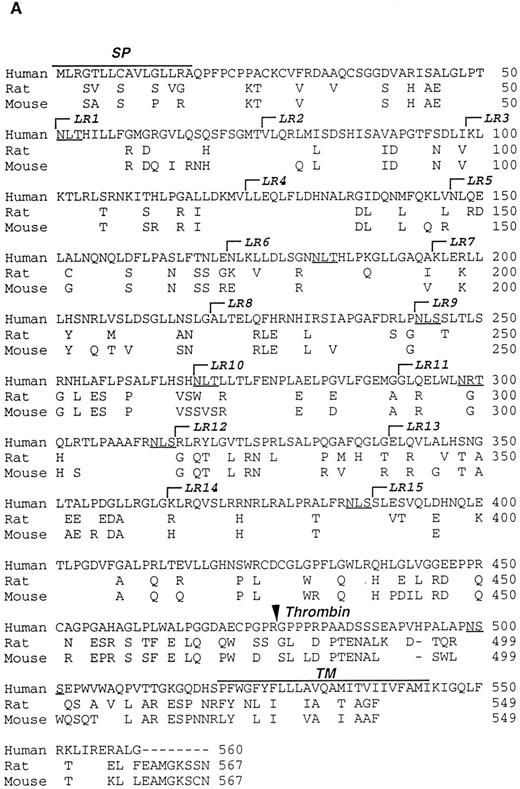
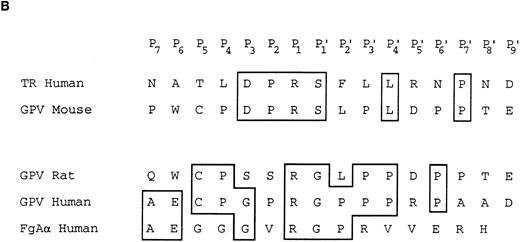
Characterization of the R and M platelet GPV proteins. (A) Comparison of the H, R, and M GPV amino acid sequences. The amino acid sequences of R and M GPV were deduced from their genes and aligned with the human sequence using CLUSTAL software. Amino acid differences with respect to the human protein are indicated. Residues encoding the signal peptide (SP) and transmembrane domain (TM) are indicated with an overline. Tandem Leu-rich (LR) motifs of 24 amino acids are marked and numbered successively. The Leu-rich boundaries have been modified as compared with Lanza16 to account for the crystal structure of another member of the Leu-rich family, the porcine RNAse inhibitor.36 Each Leu-rich motif starts at the well conserved putative β-strand segment. Potential N-glycosylation sites are underlined. A putative thrombin cleavage site is indicated by a closed arrowhead. (B) Presence of a thrombin cleavage site in R and M GPV. The putative thrombin recognition motif of M GPV is aligned with the H thrombin receptor (TR) cleavage site, while the R GPV motif is compared with the H GPV and H Aα fibrinogen (FgAα) cleavage sites. Identical residues are boxed.
Characterization of the R and M platelet GPV proteins. (A) Comparison of the H, R, and M GPV amino acid sequences. The amino acid sequences of R and M GPV were deduced from their genes and aligned with the human sequence using CLUSTAL software. Amino acid differences with respect to the human protein are indicated. Residues encoding the signal peptide (SP) and transmembrane domain (TM) are indicated with an overline. Tandem Leu-rich (LR) motifs of 24 amino acids are marked and numbered successively. The Leu-rich boundaries have been modified as compared with Lanza16 to account for the crystal structure of another member of the Leu-rich family, the porcine RNAse inhibitor.36 Each Leu-rich motif starts at the well conserved putative β-strand segment. Potential N-glycosylation sites are underlined. A putative thrombin cleavage site is indicated by a closed arrowhead. (B) Presence of a thrombin cleavage site in R and M GPV. The putative thrombin recognition motif of M GPV is aligned with the H thrombin receptor (TR) cleavage site, while the R GPV motif is compared with the H GPV and H Aα fibrinogen (FgAα) cleavage sites. Identical residues are boxed.
MALDI-TOF mass spectrometric analysis of thrombin cleavage of synthetic R, M, and H GPV peptides. Synthetic peptides corresponding to region 473-486 of R, M, and H GPV, respectively (see Materials and Methods), were incubated with saline or human α-thrombin. Thrombin treatment of rat 473-486(Y) caused loss of the nonnatural COOH-terminal tyrosine giving rise to the 473-486 peptide, which generated a thrombin cleaved 477-486 peak. M and H 473-486(Y) peptides were cleaved at position 476/477 to give peptide 477-486(Y) and a decrease of the 473-486(Y) peak. In the M spectra, the original peptide contained contaminants indicated by stars, which generated a thrombin digest peak indicated by a triangle. M/Z: observed mass/charge.
MALDI-TOF mass spectrometric analysis of thrombin cleavage of synthetic R, M, and H GPV peptides. Synthetic peptides corresponding to region 473-486 of R, M, and H GPV, respectively (see Materials and Methods), were incubated with saline or human α-thrombin. Thrombin treatment of rat 473-486(Y) caused loss of the nonnatural COOH-terminal tyrosine giving rise to the 473-486 peptide, which generated a thrombin cleaved 477-486 peak. M and H 473-486(Y) peptides were cleaved at position 476/477 to give peptide 477-486(Y) and a decrease of the 473-486(Y) peak. In the M spectra, the original peptide contained contaminants indicated by stars, which generated a thrombin digest peak indicated by a triangle. M/Z: observed mass/charge.
RESULTS
Structure of the rat and mouse GPV genes.Using a 734-nucleotide (nt) fragment from the coding sequence of human GPV, we obtained a 7.9-kb rat genomic clone and sequenced a 2.2-kb EcoRI 5′-coding fragment (λRV1) along both orientations (Fig 1A). The fragment was shown to contain 1,701 nt of coding sequence (positions 49 to 1,750), and 482 nt of 3′-untranslated sequence, but was lacking the 5′-untranslated region. PCR amplification of rat genomic DNA with antisense RP2 oligonucleotide from the 5′-end of λRV1 (nt 1,374 to 1,345, this report) and sense RP1 oligonucleotide from the human gene (nt 1,353 to 1,37516) produced a 981-nt fragment that contained 104 nt of promoter region and the entire 874 nt intron sequence. Finally, we used this amplified fragment to rescreen the rat genomic library and extended the sequence 513 nt into the 5′ promoter region. The λRV1 fragment was used to clone the mouse GPV gene from a mouse genomic library in the λ Dash vector. A 3.5-kb EcoRI fragment (λMV1) from a 22.5-kb positive clone contained 511 nt of 5′-untranslated and promoter region, 894 nt of intron, 1,704 nt corresponding to the entire coding region (positions 1,408 to 3,111), and 472 nt of 3′-untranslated region (Fig 1A).
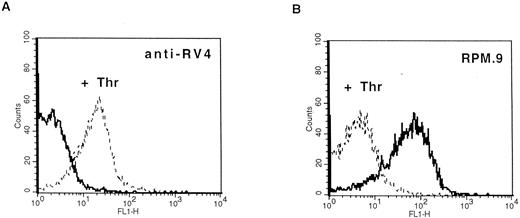
Detection of thrombin-activated rat platelets by FACS analysis with the anti-RV4 polyclonal antibody and MoAb RPM.9. Washed rat platelets (2 × 105) were incubated with 2 μg of anti-RV4 followed by FITC-labeled goat antirabbit antibody (A) or with 2 μg of MoAb RPM.9 followed by FITC-labeled goat antimouse antibody (B) after treatment with 5 U/mL thrombin (dashed line) or without treatment (solid line). (A) Anti-RV4 did not bind to resting platelets and gave a fluorescence signal equivalent to that of a nonimmune serum (data not shown). Treatment with thrombin resulted in a positive shift in fluorescence suggesting that anti-RV4 recognizes the new NH2-terminus generated by cleavage of rat GPV. (B) On the contrary, MoAb RPM.9 positively labeled resting rat platelets, while thrombin treatment shifted the fluorescence signal to negative values. Thus, MoAb RPM.9 appears to bind to a fragment NH2-terminal to the thrombin cleavage site, which is released from thrombin activated platelets.
Detection of thrombin-activated rat platelets by FACS analysis with the anti-RV4 polyclonal antibody and MoAb RPM.9. Washed rat platelets (2 × 105) were incubated with 2 μg of anti-RV4 followed by FITC-labeled goat antirabbit antibody (A) or with 2 μg of MoAb RPM.9 followed by FITC-labeled goat antimouse antibody (B) after treatment with 5 U/mL thrombin (dashed line) or without treatment (solid line). (A) Anti-RV4 did not bind to resting platelets and gave a fluorescence signal equivalent to that of a nonimmune serum (data not shown). Treatment with thrombin resulted in a positive shift in fluorescence suggesting that anti-RV4 recognizes the new NH2-terminus generated by cleavage of rat GPV. (B) On the contrary, MoAb RPM.9 positively labeled resting rat platelets, while thrombin treatment shifted the fluorescence signal to negative values. Thus, MoAb RPM.9 appears to bind to a fragment NH2-terminal to the thrombin cleavage site, which is released from thrombin activated platelets.
The structures of the rat and mouse GPV genes are well conserved relative to the human gene, a similar size intron interrupts a small 5′-untranslated region and the entire coding sequence is contained within the second exon. Nucleotide sequence identity between the human and rodent genes is 79% in the coding region, while there is only about 60% identity in the intron (Table 1). Alignment of the first 500 nt preceding the intron (Fig 1B) shows extensive homology between the three species with 67% identity in the first 220 nt. This region contains a putative transcription initiation site located 29 nt upstream of the donor splice site. This site is 100% conserved between the three species, matches the consensus sequence for initiation found in the mouse terminal transferase gene, and is highly homologous to sites present in other megakaryocyte expressed genes such as the αIIb and α2 integrins, the PF4, and the GPIX genes (Fig 1C).14,18,34,35 In addition, the region upstream of this initiation site contains several conserved consensus recognition sites for nuclear transcription factors of the hematopoietic lineage.17
Amino acid sequences of rat and mouse GPV.Rat and mouse GPV contain 567 amino acids, as compared with 560 amino acids for human GPV (Fig 2A). All three proteins possess an NH2-terminal 16 amino acid signal peptide and a COOH-terminal 25 amino acid transmembrane domain. Rat and mouse GPV contain eight additional amino acids in the intracellular region and lack one amino acid in the extracellular domain at position 493. This gives a mature protein of 551 amino acids for rat and mouse GPV as compared with 544 amino acids for human GPV. The extracellular domains of rat and mouse GPV display like the human protein 15 tandem repeats of 24 amino acids enriched in leucines with very high conservation for leucines and for one asparagine at positions that are also conserved within the Leu-rich family of proteins.36 Similar to human GPV, sequences in the vicinity of residue 476 in rat and mouse GPV contain a potential thrombin cleavage site (Fig 2B).
Thrombin cleavage of synthetic rat, mouse, and human GPV peptides.To test for functionality of the putative thrombin cleavage sites, we synthesized peptides comprising residues 473 to 486 of rat, mouse, and human GPV (see Materials and Methods). These peptides were incubated with purified human α-thrombin and cleavage was assessed by analysis of the reaction products by MALDI-TOF mass spectrometry (Fig 3) in comparison with control 473-486 and 477-486 peptides. Mass spectrometric analysis demonstrated precise cleavage by thrombin between positions 476 and 477 for all three species. In the rat RV1 peptide, unexplained cleavage occurred at a nonnatural tyrosine at position 486, whereas the mouse MV1 peptide was not cleaved at this position. Control incubations without enzyme for 24 hours at 37°C resulted in no apparent cleavage of the peptides, and cleavage at position 476 did not occur with other proteases such as leukocyte elastase or cathepsin G (data not shown). Parallel experiments using HPLC separation confirmed the cleavage observed by mass spectrometry (data not shown). These experiments thus demonstrate the presence of a functional thrombin cleavage site at a position conserved in human, rat, and mouse GPV.
Production of antibodies specific for rat GPV and characterization of the protein.A synthetic peptide (RV4) between residues 477 and 490 corresponding to the new NH2-terminus generated after thrombin cleavage was coupled to KLH and used to raise polyclonal antibodies in rabbits. Immunoblotting analysis showed that anti-RV4 specifically labeled an 88-kD band from rat platelet proteins and purified rat GPV, and recognized a 14-kD band from thrombin treated and deglycosylated platelet proteins (data not shown). This band probably corresponds to the COOH-terminal, membrane attached fragment generated after thrombin cleavage. To test for reactivity of anti-RV4 against native GPV, FACS analysis was performed on washed rat platelets (Fig 4). Unstimulated platelets were not recognized by anti-RV4 contrary to thrombin-treated platelets, which gave a strong positive signal. This suggested that thrombin cleavage of the 476-477 bond unmasked the epitope recognized by the anti-RV4 antibody. The increase in fluorescence was specific for thrombin and was not observed when rat platelets were stimulated with other agonists such as ADP or collagen. In addition, anti-RV4 was specific for rat platelets and did not recognize unstimulated or thrombin-stimulated platelets from mice or humans (data not shown).
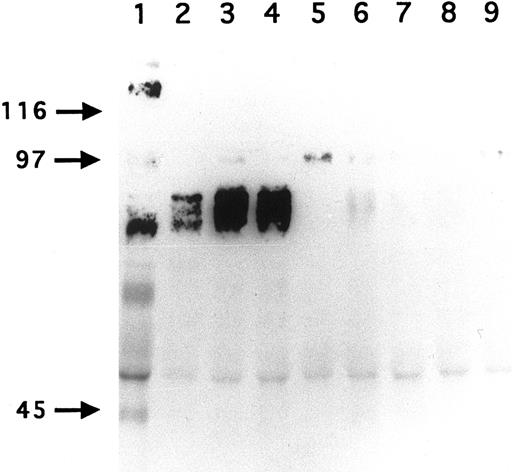
Immunoprecipitation of GPV from biotinylated H and R platelet proteins. H (lane 1) and R (lanes 2 to 9) platelets were surface labeled with biotin and treated with saline (lanes 1 to 5) or with 5 U/mL thrombin (lanes 6 to 9). Platelets were washed, lysed with Triton X-100, and the proteins immunoprecipitated with MoAb V.1 against human GPV (lane 1), MoAbs RPM.4 (lanes 2 and 6), RPM.9 (lanes 3 and 7), and RPM.11 (lanes 4 and 8) against R GPV, and MoAb RPM.10 against R GPIIb-IIIa (lanes 5 and 9). MoAb V.1 immunoprecipitated a 82-kD band from human resting platelets (lane 1). MoAbs RPM.4, RPM.9, and RPM.11 immunoprecipitated a broad band centered at 88 kD from R unstimulated platelets (lanes 2 to 4). This band disappeared from the surface of thrombin-stimulated platelets (lanes 6 to 8). These results confirm that R GPV is specifically cleaved by thrombin and that the epitopes for the R GPV MoAbs are located NH2-terminal from the cleavage site. Under these conditions, RPM.10 gave a weak signal at 100 kD (lane 5) corresponding to GPIIIa and no labeling of GPIIb.
Immunoprecipitation of GPV from biotinylated H and R platelet proteins. H (lane 1) and R (lanes 2 to 9) platelets were surface labeled with biotin and treated with saline (lanes 1 to 5) or with 5 U/mL thrombin (lanes 6 to 9). Platelets were washed, lysed with Triton X-100, and the proteins immunoprecipitated with MoAb V.1 against human GPV (lane 1), MoAbs RPM.4 (lanes 2 and 6), RPM.9 (lanes 3 and 7), and RPM.11 (lanes 4 and 8) against R GPV, and MoAb RPM.10 against R GPIIb-IIIa (lanes 5 and 9). MoAb V.1 immunoprecipitated a 82-kD band from human resting platelets (lane 1). MoAbs RPM.4, RPM.9, and RPM.11 immunoprecipitated a broad band centered at 88 kD from R unstimulated platelets (lanes 2 to 4). This band disappeared from the surface of thrombin-stimulated platelets (lanes 6 to 8). These results confirm that R GPV is specifically cleaved by thrombin and that the epitopes for the R GPV MoAbs are located NH2-terminal from the cleavage site. Under these conditions, RPM.10 gave a weak signal at 100 kD (lane 5) corresponding to GPIIIa and no labeling of GPIIb.
To develop reagents specific for rat platelet and to further characterize rat GPV, we raised MoAbs in mice immunized with rat platelets. Three GPV-specific antibodies, RPM.4, RPM.9, and RPM.11, were detected by FACS screening on rat platelets treated or untreated by thrombin. A representative histogram for antibody RPM.9 is shown in Fig 4 displaying strong reactivity against unstimulated platelets and a decrease in fluorescence on thrombin treatment. This suggested reactivity against the extracellular domain of rat GPV released after thrombin cleavage. Immunoprecipitation experiments using biotinylated resting rat platelets (Fig 5) showed a broad 88-kD band characteristic of highly glycosylated GPV. Treatment of rat platelets with 5 U/mL human thrombin led to a loss of reactivity towards the antibodies (Fig 5) and to liberation of a soluble 70-kD fragment (data not shown) thus confirming that rat GPV is cleaved from the platelet surface and that the epitopes for the three antibodies are located NH2-terminal from the cleavage site. The antibodies are specific for rat platelets, as none of them reacted with human platelets (data not shown).
DISCUSSION
This report describes the isolation and characterization of the rat and mouse platelet GPV genes and compares their structures with the human counterpart. The identity of the rat and mouse genes was established by their high homology with human GPV at the DNA and amino acid levels and for rat GPV by the development of species specific antibodies. The main findings of this study are: (1) very similar genomic structure of rat, mouse and human GPV; (2) a well-conserved promoter region with megakaryocyte specific features; (3) high sequence identity at the amino acid level especially in the Leu-rich region; and (4) maintenance of a functional thrombin cleavage site in rat and mouse GPV. Finally, we developed antibodies specific for rat GPV, which are able to discriminate between resting and thrombin-activated platelets.
Human GPV is characterized, like the other platelet members of the Leu-rich family of proteins, GPIbα, GPIbβ, and GPIX, by a very simple genomic stucture.10,12,14,16 In this study, we show that this simple organization also applies to the GPV gene from other species. Comparison of the two rodent genes with their human counterpart showed a similar intron-exon distribution, the second exon containing the entire coding sequence. A putative transcription start site is located in all three species 29 nt from the gt donor site, and is most probably functional as it conforms to the start site in other platelet-specific genes and matches the human megakaryocyte GPV cDNA end obtained by the 5′ RACE technique.16 The start site previously reported at position 1,294 of the human gene16 might not be the major functional site, as it is not conserved in rat and mouse.
Comparison of the promoter regions showed good candidates for functional cis-acting elements, notably two GATA and two ets sites in the region -46 to -113, which are identical in the three species. GATA and ets motifs are also found in the human and mouse GPIIb gene promoters, where their assocation contributes to the megakaryocyte specific expression of GPIIb.19 Their conservation in different species would support their role in controlling the transcription of GPV. The mouse and rat promoters lack the two TATA boxes found in the human gene. This lack of conservation and the remoteness of the TATA motifs from the Cap site in human GPV raises questions as to their functional importance, although functional TATA boxes of the human PF4 and tPA genes were equally found to be absent in the respective rodent genes.37,38 Two cis-acting elements potentially involved in the megakaryocyte specific expression of GPV are only found in the rodent promoters. First, a PMS-E site in mouse and rat, a site that was identified in the human GPIIb promoter and shown to bind megakaryocyte specific factors.18 Second, an NFE-2 site in the rat gene, a site that was recently shown by gene disruption in mice to play an essential role in megakaryocyte maturation.39
Before this report, no information was available concerning nonhuman sequences of the four subunits forming the GPIb-V–IX complex. The rat and mouse GPV proteins have an overall organization very similar to that of human GPV, the only major difference being the addition of eight amino acids in the intracellular region. Alignment of the protein sequences gave 86% identity between rat and mouse GPV and 70% identity between rat or mouse and human GPV. A similar degree of conservation (≈75%) is observed for other rodent platelet proteins like P-selectin or the thrombin receptor.40,41 The Leu-rich and transmembrane domains are the most conserved regions, whereas there is less conservation in the sequence connecting the Leu-rich segment to the membrane. The predicted molecular weights for the mature unglycosylated proteins were 63.3 kD for rat and 63.5 kD for mouse GPV. These sizes are slightly larger than unglycosylated human GPV (59.2 kD) mainly because of the larger intracellular segment. The eight cysteines found in human GPV are exactly conserved in rat and mouse and from the Cys bonds assignment for human GPIbα,42 we can assume that GPV in the three species has its Leu-rich domain flanked on each side by a double Cys loop structure.
Rat and mouse GPV both have 15 Leu-rich repeats like human GPV, a number that might be critical to maintain correct conformation of the protein for its association with GPIb-IX or for some potential binding function. The repeats are in good agreement with the 1-XLXXLXLXXNXLXXLPXXLFXXLX-24 consensus sequence derived from human GPIbα and GPV.9,16 This similar distribution of Leu residues suggests that the three proteins would fold into identical three-dimensional structures. The only known x-ray structure of a Leu-rich family member is that of RNAse inhibitor36 where each repeat of 28 or 29 amino acids is formed by a right-handed β-α structural unit giving a horseshoe-shaped superhelical structure. Modeling of the repeats containing 20 to 26 amino acids43 predicts a very similar structure with a β-strand segment of 10 amino acids connected to a shorter α-helical segment. To date, the exact role of the Leu-rich domains for the proteins forming the GPIb-V–IX complex is not fully understood. They could provide a structural backbone allowing ligand binding or subunits association. In favor of this hypothesis, several mutations have been described in Leu-rich regions of GPIbα and GPIX in patients with the Bernard-Soulier bleeding disorder, which lead to defective von Willebrand factor binding and decreased stability or association of the GPIb-V–IX complex.44
Human GPV is unique among the major platelet surface glycoproteins by being cleaved by thrombin during platelet activation. The present study clearly demonstrates the existence of a functional thrombin recognition site in GPV from other species. Maintenance of this site across the three species is remarkable, as it occurs in the least conserved region of the protein and this would favor a role for GPV cleavage in platelet physiology. One surprising finding was that the mouse cleavage site has diverged from those of the human and more closely related rat proteins to now resemble that of the human thrombin receptor.26 These two sites display 100% identity from residues P3 to P′1 with an unfavorable Ser at P′1.45 Despite this unfavorable site, a mouse GPV peptide was cleaved as efficiently as the corresponding human peptide.
Availability of the genes coding for mouse and rat GPV and of specific reagents for rat GPV should facilitate future ex vivo and in vivo functional studies in animal models. Thus we raised rat-specific antibodies and characterized the biochemical properties of the rat GPV protein. The rat protein is slightly larger than its human counterpart, 88 kD as compared with 82 kD, has a similar size heterogeneity due to complex glycosylation, and is similarly released by thrombin cleavage. The two types of antibodies raised can be used in thrombosis models either to detect thrombin-liberated GPV in plasma (MoAbs RPM.4, RPM.9, and RPM.11) or to detect thrombin-activated platelets (anti-RV4). GPV could, in fact, offer advantages over known markers of platelet activation such as GPIIb-IIIa, PF4, or P-selectin, because it is restricted to platelets and would be a direct indicator of the presence of active thrombin. In view of the very similar properties of rat and human GPV, it is assumed that results obtained using rat models could be applicable to humans.
Knowledge of the mouse gene also opens the possibility of performing gene inactivation experiments to unravel the exact physiological role of GPV. Transfection experiments certainly indicate a role of GPV in the assembly and expression of the GPIb-V–IX complex21,22 and suggest that certain cases of Bernard-Soulier disease could be explained by GPV defects. A second possible consequence of a lack of GPV would be to inhibit platelet responses to thrombin.46 One recent report that disruption of the thrombin receptor had no effect on platelet responses to thrombin47 supports the presence of a second thrombin receptor. It will now be possible to directly assess the involvement of GPV after gene inactivation in the thrombin receptor-deficient strain.
In conclusion, the data presented here should prove useful in understanding the regulation of the megakaryocyte-specific expression of genes and also for further studies aimed at defining the function of the poorly characterized GPV protein.
ACKNOWLEDGMENT
The authors thank Monique Freund for her help in raising antibodies against rat GPV and Juliette N. Mulvihill for correcting the English.
Supported by a postdoctoral fellowship from Eli Lilly (Indianapolis, IN) to D.O.A.
Presented in part at the Fifteenth meeting of the International Society of Thrombosis and Haemostasis, Jerusalem, Israel, June 11–16, 1995, and published in abstract form in Thromb Haemost 73:1196, 1995.
Address reprints requests to François Lanza, PhD, INSERM Unité U.311, Etablissement de Transfusion Sanguine de Strasbourg, 10 rue Spielmann, BP36, 67065 Strasbourg Cedex, France.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal