Abstract
The adhesion molecule BEN/SC1/DM-GRASP (BEN) is a marker in the developing chicken nervous system that is also expressed on the surface of embryonic and adult hematopoietic cells such as immature thymocytes, myeloid progenitors, and erythroid progenitors. F84.1 and KG-CAM, two monoclonal antibodies to rat neuronal glycoproteins with similarity to BEN, cross-react with an antigen on rat hematopoietic progenitors, but F84.1 only also recognizes human blood cell progenitors. We have defined the antigen recognized by F84.1 as the hematopoietic cell antigen (HCA). HCA expression was detected on 40% to 70% of CD34+ fetal and adult bone marrow cells and mobilized peripheral blood cells. Precursor cell activity for long-term in vitro bone marrow cell culture was confined to the subset of CD34+ cells that coexpress HCA. HCA is expressed by the most primitive subsets of CD34+ cells, including all rhodamine 123lo, Thy-1+, and CD38−/lo CD34+ adult bone marrow cells. HCA was also detected on myeloid progenitors but not on early B-cell progenitors. We also describe here the cloning and characterization of cDNAs encoding two variants of the human HCA antigen (huHCA-1 and huHCA-2) and of a cDNA clone encoding rat HCA (raHCA). The deduced amino acid sequences of huHCA and raHCA are homologous to that of chicken BEN. Recombinant proteins produced from either human or rat HCA cDNAs were recognized by F84.1, whereas rat HCA but not human HCA was recognized by antirat KG-CAM. Expression of either form of huHCA in CHO cells conferred homophilic adhesion that could be competed with soluble recombinant huHCA-Fc. The molecular cloning of HCA and the availability of recombinant HCA should permit further evaluation of its role in human and rodent hematopoiesis.
CELL SURFACE PROTEINS common to both hematopoietic and nervous systems have been described, including cytokine receptors and adhesion molecules (eg, c-kit, integrins, N-CAM, and Thy-1), some of which are also expressed on hematopoietic stem/progenitor cells (reviewed in Knapp et al1 ). The adhesion molecule BEN, also discovered independently as SC1 and DM-GRASP, was originally described as a marker in the developing central and peripheral chicken nervous systems of subsets of motor and sensory neurons, during phases of axonogenesis, neurite elongation, axonal fasciculation, and synapse formation.2-4 BEN expression was also detected in the chicken embryo at the surface of paraaortic early hematopoietic stem cells (HSCs),5 on adult bone marrow (BM) myeloerythroid progenitors, on immature double-positive thymocytes, and on activated T lymphocytes.6,7 The monoclonal antibody (MoAb) F84.1 recognizes a 90- to 105-kD cell surface antigen on rat neurons that has similar amino-terminal sequence to chicken BEN8 and cross-reacts with an antigen on a subfraction of human BM cells.9 We have defined the human antigen recognized by F84.1 as the hematopoietic cell antigen (huHCA).
In this study, we describe the characterization of HCA+ cells in human fetal and adult hematopoietic tissues and show that HCA is expressed by the most primitive hematopoietic cells. We also report the cloning and characterization of cDNAs encoding two variants of the human HCA antigen (huHCA-1 and huHCA-2) and of a cDNA clone encoding rat HCA (raHCA). The deduced amino acid sequences of huHCA and raHCA are homologous to that of the chicken BEN adhesion molecule. Antigenic differences between human and rat HCA were observed using a second MoAb, 11-59 (antirat KG-CAM), that recognizes raHCA but not huHCA. Expression of huHCA in CHO cells conferred homophilic adhesion that could be competed with a soluble recombinant huHCA-Fc fusion protein but not by F84.1. These data suggest a role for HCA in the adhesion of stem cells and progenitor cells to themselves or to other HCA+ supportive cells in the microenvironment of blood-forming tissues.
MATERIALS AND METHODS
Human tissues.Human embryonic and early fetal tissues were obtained from voluntary or therapeutic abortions performed in compliance with the French legislation. Developmental stages (ie, postconception) were estimated from menstrual history and confirmed by anatomic criteria; 18- to 20-week fetal bones were obtained with informed consent after elective abortions. Adult BM (ABM) was aspirated from the posterior iliac crest of healthy adult volunteers under IRB approval at the Scripp's Clinic (San Diego, CA). Leukapheresed mobilized peripheral blood (MPB) samples were obtained from multiple myeloma patients treated (cyclophosphamide at 6 g/m2 at day 0, followed by the administration of granulocyte-macrophage colony-stimulating factor [GM-CSF ] at 0.25 mg/m2) at University of Arkansas Medical Center after informed consent was received. Fetal BM (FBM) cells were flushed from split long bones and ficolled to obtain low-density mononuclear cells (Lymphoprep; Nycomed Pharmacia, Oslo, Norway). Low-density (<1.077 g/mL) mononuclear ABM cells were isolated by using Ficoll-hypaque (Pharmacia, Piscataway, NJ). CD34+ ABM cells were positively selected before fluorescence-activated cell sorter (FACS) isolation by using affinity columns (Cellpro, Bothell, WA) or paramagnetic beads (CD34 selection kit; Miltenyi, Sunnyvale, CA). Red blood cells in leukapheresis MPB samples were lysed with 0.83% ammonium chloride lysis buffer for 5 to 10 minutes before antibody staining.
Immunohistochemistry.Freshly dissected embryonic and fetal human tissues were fixed in 4% paraformaldehyde and then impregnated and included in gelatin/saccharose medium before deep freezing in isopentane vapors, as described previously.10 The F84.1 antibody, under hybridoma supernatant form (mouse IgG1) and diluted 1:20 in PBT (phosphate-buffered saline [PBS] with 0.25% Triton), was applied overnight on cryostat sections that were then rinsed three times for 10 minutes in PBT. Immune reaction was shown using the ABC biotin-streptavidin kit (Vector, Burlingame, CA) according to the manufacturer's instructions.
Rhodamine 123 staining.The staining procedure for Rhodamine 123 (Rh123) was described previously11 12 and modified as follows. A 1 mg/mL stock solution of Rh123 (Molecular Probes, Eugene, OR) was prepared in ethanol and stored at −20°C in the dark just before use. ABM, FBM, or MPB cells were resuspended at 106/mL or less in buffer (Hanks' Balanced Salt Solution [HBSS], RPMI with HEPES or PBS) containing 2% fetal calf serum (FCS) and incubated with 0.1 mg/mL of Rh123 dye for 30 minutes at 37°C. The cells were washed and incubated at 37°C for 40 minutes to allow efflux of the Rh123 dye, washed again, and stained with antibodies. Rh123 fluorescence was analyzed by flow cytometry in the fluorescein isothiocyanate (FITC) channel after setting compensation between FITC and phycoerythrin (PE) channels.
Antibodies.The MoAb F84.18 was obtained from Dr W. Stallcup (La Jolla Cancer Foundation, La Jolla, CA). The MoAb to KG-CAM, 11-59,13 was obtained from Dr E. Geisert Jr (University of Tennessee, Memphis, TN). PE- and FITC-conjugated goat antimouse IgG1 antibodies were purchased from Caltag (South San Francisco, CA). The CD34 antigen was detected by staining cells with anti-CD34 (Tuk3, mouse IgG3; from Dr A. Ziegler, University of Berlin, Berlin, Germany) and Texas Red-conjugated goat antimouse IgG3 (Southern Biotechnology Associates, Birmingham, AL). Allophycocyanin (APC)- and sulfarhodamine-conjugated Fab′2 Tuk-3 reagents were prepared at SyStemix. hHCA expression was detected by staining cells with F84.1 MoAb (mouse IgG1) and PE-conjugated goat antimouse IgG1 (Caltag). The following PE-conjugated antilineage marker antibodies were used to define lineage-negative (Lin−) populations: anti-CD19 for the B-cell lineage, anti-CD2 for the T-cell lineage, anti-CD14 for myelomonocytic lineages (Becton Dickinson, San Jose, CA), and anti-glycophorin A (AMAC, Westbrook, ME) for the erythroid lineage. The isotype-matched negative control antibodies were used to delineate gated populations. The cells were incubated for 20 minutes on ice for each step. After the final wash, cells were resuspended in HBSS containing 1 mg/mL propidium iodide (PI). Labeled cells were analyzed and sorted with a dual-laser FACS (Becton Dickinson Immunocytometry Systems, San Jose, CA) at the SyStemix FACS facility. Dead cells were excluded from analysis by their PI staining characteristics. After sorting, CD34+ populations were checked by FACS reanalysis. In some cases, hematopoietic cells were stained with Rh123 dye before MoAb staining.
In vitro long-term cobblestone area-forming cell (CAFC) assays.Sorted cells were cultured on a pre-established monolayer of the SyS-1 mouse stromal cell line in 96-well flat-bottom plates as described14-16 in the presence of 50 ng/mL leukemia inhibitory factor (LIF ) and 10 ng/mL interleukin-6 (IL-6). Cultures were fed weekly by replacing 50% of the medium consisting of 50% Iscove's modified Dulbecco's medium (IMDM; JRH Bioscience, Lenexa, KS), 50% RPMI with 10% FCS (Hyclone Laboratories, Logan, UT), 4 × 10−5 mol/L 2-mercaptoethanol, 100 U/mL penicillin-streptomycin, and 4 mmol/L glutamine (JRH Bioscience), 50 ng/mL LIF, and 10 ng/mL human IL-6. To perform CAFC assays (measured at weeks 3 through 8), HCA+ Rh123lo (1 and 3 cells/well), HCA+ Rh123mid (1 and 3 cells/well), HCA+ Rh123hi (5 and 15 cells/well), and HCA− Rh123hi (5 and 15 cells/well) were sorted with the automated cell deposition unit (ACDU) into 96-well plates preseeded with SyS-1 stromal cells, with one or two 96-well plates per cell concentration being used for each population. Based on two independent experiments, linear regression analysis of the proportion of negative wells at each concentration was used to determine the frequency of CAFC at 3, 4, 5, and 6 to 8 weeks. The CAFCs were harvested 6 to 8 weeks after plating and analyzed for CD34, CD33, and CD19 expression by FACS.
Molecular cloning of HCA.Total cellular RNA was extracted from 5 × 105 CD34+ cells using the phenol/guanidinium isothiocyanate method.17 Oligo dT-primed first-strand cDNA synthesis was performed using a kit purchased from GIBCO (Gaithersberg, MD). Candidate HCA cDNA clones were amplified using the polymerase chain reaction (PCR) using degenerate 5′-oligonucleotide primers based on the published amino-terminal sequences of either the F84.1 antigen8 (HCA51) or the KG-CAM antigen13 (HCA52) and using a degenerate 3′-oligonucleotide primer based on the consensus cytoplasmic domain sequence of chicken BEN and its homologue in fish, neurolin18 (HCA31). The PCR reaction was performed for 40 cycles of the following conditions: 94°C for 45 seconds, 50°C for 35 seconds, and 72°C for 90 seconds, PCR products were subcloned into pCRII (Invitrogen, La Jolla, CA) before sequencing on a Model 373 DNA Sequencer using reagents purchased from Applied Biosystems (Foster City, CA). Complete 5′ and 3′ sequences of candidate HCA clones were derived by PCR-RACE of CD34+ cell cDNA using a kit from GIBCO and by screening a human BM cDNA library (Clontech, Palo Alto, CA) with 32P-labeled HCA cDNA. Rat HCA cDNAs were derived from a rat B49 cell cDNA library (Z.Y., unpublished data) by screening with a 32P-labeled human HCA cDNA fragment prepared by random priming.19 All other DNA manipulations were performed using conventional procedures.20
Immunochemical characterization of huHCA-Fc and raHCA-Fc fusion proteins.Soluble forms of huHCA and raHCA were produced by directional cloning of the extracellular domain of the coding regions into the mammalian expression vector pCD5neg-1.21 To produce the recombinant HCA-2-Fc fusion protein, DNA fragments corresponding to the extracellular domain of huHCA-2 were generated by PCR using synthetic oligonucleotides and ligated to pCD5neg-1.21 The resulting constructs contain fused sequences encoding the CD5 signal sequence and the huHCA or raHCA extracellular domain-IgG1Fc fusion gene in tandem and direct the secretion of soluble huHCA-Fc and raHCA-Fc, respectively, fusion proteins. COS-7 cells were transfected with these constructed vectors, and media were harvested. Protein A-sepharose immunochromatography was used to purify the fusion protein from the media.
Enzyme-linked immunosorbent assays (ELISAs) were performed by coating huHCA-Fc, raHCA-Fc, or CD14-Fc onto an Immulon 4 ELISA plate (Dynatech, Chantilly, VA) at 10 μg/mL, which was then saturated using 2% bovine serum albumin (BSA), 0.05% Tween-20 in PBS, pH 7.2. Samples containing MoAbs F84.1, 11-59, or an IgG1 isotype control were added. After MoAb incubation for 1 hour at room temperature (RT), the plates were incubated with alkaline phosphatase-goat antimouse Ig for 1 hour at RT. The plates were washed and developed using p-nitrophenylphosphate and read at 405 nm using a Molecular Devices ELISA reader (Sunnyvale, CA).
Expression of HCA cDNAs in CHO cells.Full-length HCA cDNAs were constructed by ligation of appropriate restriction fragments to the mammalian expression vector pSRa.22 To generate stable HCA expression cell lines, CHO cells were cotransfected with pSRa-HCA and pSV2-neo (5:1 ratio) by a liposome-mediated transfection procedure (GIBCO). The transfected CHO cells were selected with 0.8 mg/mL G418.
Immunoprecipitation assays.Transfected cells were washed once in PBS and starved in methionine- and cysteine-free Dulbecco's modified Eagle's medium (DMEM) containing 5% dialyzed FCS for 1 hour at 37°C. Cells (2 × 106/mL) were then metabolically labeled with 35S-methionine and 35S-cysteine (Amersham; Arlington Heights, IL; 800 μCi/mL) for 4 hours. The labeled CHO cells were lysed in 0.1% Triton X-100/PBS. Lysates were incubated with either MoAb F84.1 or 11-59 at 4°C for 45 minutes. Magnetic beads conjugated with antimouse IgG1 antibody were mixed with the antibody-lysate mixture for 35 minutes and then isolated using a magnet. The beads were washed three times with 0.1% Triton X-100/PBS. Bound antigens were released by heating the beads at 80°C in 0.1% sodium dodecyl sulfate (SDS) sample buffer and loaded onto a 4% to 20% SDS gel. For production of recombinant HCA-1-Fc fusion proteins, the supernatant of transfected COS7 cells was collected after 72 hours and the HCA-1-Fc was isolated using protein A-Sepharose beads (Pharmacia).
Adhesion assays.Subconfluent monolayers of HCA-expressing CHO cells or parental CHO cells were washed with Ca2+- and Mg2+- free PBS twice and then detached from the dish with 0.3% EDTA/PBS (pH 7.4). The cells were then collected and a single-cell suspension was prepared in DMEM containing 0.3% EDTA, 1 mg/mL chondroitin sulphate (Sigma, St Louis, MO), 0.0002% sodium azide, and 2% BSA at 1 × 106 cells/mL. The cells were kept on ice to prevent aggregation. Cell aggregation assays were performed by incubating 1-mL aliquots of the cell suspension at 25°C with or without agitation. Cell clumping was assessed visually after 30 minutes. To confirm that the cell aggregation was HCA-mediated, 40 μg/mL purified HCA-1-Fc fusion protein or human IgG1 was added to the cells for 30 minutes on ice before starting the assay.
RNA blots.Filters preloaded with mRNA from different human fetal and adult tissues were purchased from Clontech. A 1.8-kb HCA cDNA probe was prepared by random priming.19 RNA filter hybridizations were performed overnight at 65°C in 0.5 mol/L NaH2PO4 (pH 7.2), 7% SDS, 1 mmol/L EDTA, 1% BSA.20 Blots were washed at high stringency (0.1× standard saline citrate [SSC; 1× SSC is 150 mmol/L NaCl, 15 mmol/L sodium citrate, pH 7.0], 1 mmol/L NaH2PO4 , 0.5% SDS at 60°C).
RESULTS
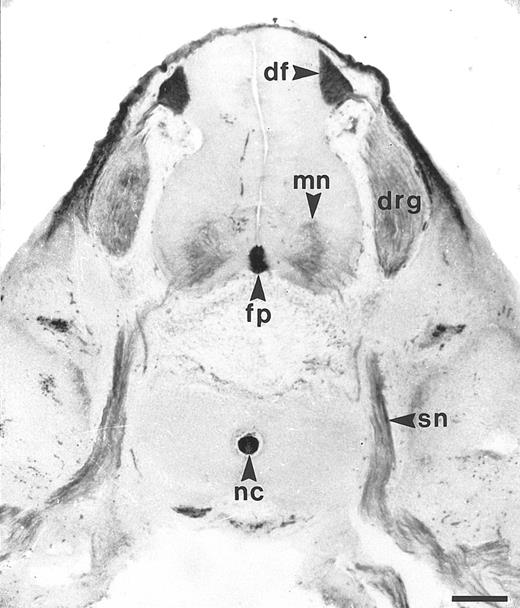
Similar to the chicken BEN antigen, HCA is expressed by subsets of human developing neurons and hematopoietic cells.The reactivity of the F84-1 antibody was assayed on transverse trunk sections of 7-week human embryos. At that stage, the majority of motoneurons in the neural tube have been born and have started extending axons to form the spinal nerves; both cell bodies and axons then display strong F84.1 reactivity (Fig 1). At the midline of the neural tube, the floor plate is also intensely stained, as are the cell bodies and axons of the dorsal root ganglion (DRG) neurons, which arise from the neural crest (Fig 1). The central projections of these axons constitute the dorsal funiculus, which is also brightly labeled by F84.1. Nonneuronal tissues such as the notochord and the mesenchyme overlying the DRGs and spinal cord are also strongly reactive. Therefore, HCA is expressed on developing human spinal cord neurons during phases of axonal elongation and fasciculation (Karagogeos et al, in press) and thus exhibits a distribution pattern very similar to that of BEN in the nervous system of the chicken embryo.2-4,6 10
Expression of F84.1/HCA in the developing human spinal cord. The developing human spinal cord was studied using F84.1 MoAb on transverse sections of human embryos, at the trunk level, a 7 weeks postconception. The majority of the motoneurons (mn) of the neural tube have been born and have started to extend axons to form the spinal nerves (sn). Both their cell bodies and axons display strong F84.1 reactivity. At the midline of the neural tube, the floor plate (fp) is also intensely stained. Cell bodies and axons of the DRG neurons that arise from the neural crest also exhibit a strong expression of F84.1 epitope. The central projections of these axons form the dorsal funiculus (df ) in the spinal cord, which is intensely labeled. Nonneuronal tissues such as the notochord and surface ectoderm are also strongly reactive.
Expression of F84.1/HCA in the developing human spinal cord. The developing human spinal cord was studied using F84.1 MoAb on transverse sections of human embryos, at the trunk level, a 7 weeks postconception. The majority of the motoneurons (mn) of the neural tube have been born and have started to extend axons to form the spinal nerves (sn). Both their cell bodies and axons display strong F84.1 reactivity. At the midline of the neural tube, the floor plate (fp) is also intensely stained. Cell bodies and axons of the DRG neurons that arise from the neural crest also exhibit a strong expression of F84.1 epitope. The central projections of these axons form the dorsal funiculus (df ) in the spinal cord, which is intensely labeled. Nonneuronal tissues such as the notochord and surface ectoderm are also strongly reactive.
With respect to the reported coexpression of the chicken neuron-associated BEN antigen by immature hematopoietic cells in the embryonic and posthatching life,5 our initial observation of F84.1 reactivity in the human embryonic spinal cord prompted the analysis of human ABM mononuclear cell suspensions, inside which about 30% cells were found to be HCA+ by single-color FACS analysis (Fig 2A). Expression of HCA-like antigens on hematopoietic cells was further examined by staining with the 11-59 MoAb that recognizes a rat antigen, KG-CAM, with an amino-terminal sequence similar to that of BEN.8 13 F84.1 recognizes a subset of both human and rat BM cells (Fig 2A and C), whereas 11-59 stains rat cells but not human cells (Fig 2B and D).
Reactivity of binding of the MoAbs F84.1 and 11-59 with human and rat hematopoietic cells. Human (A and B) or rat (C and D) mononuclear BM cells were stained with F84.1 (shaded in A and C), 11-59 (shaded in B and D), or an isotype-matched control antibody (not shaded, all panels) and analyzed by FACS.
Reactivity of binding of the MoAbs F84.1 and 11-59 with human and rat hematopoietic cells. Human (A and B) or rat (C and D) mononuclear BM cells were stained with F84.1 (shaded in A and C), 11-59 (shaded in B and D), or an isotype-matched control antibody (not shaded, all panels) and analyzed by FACS.
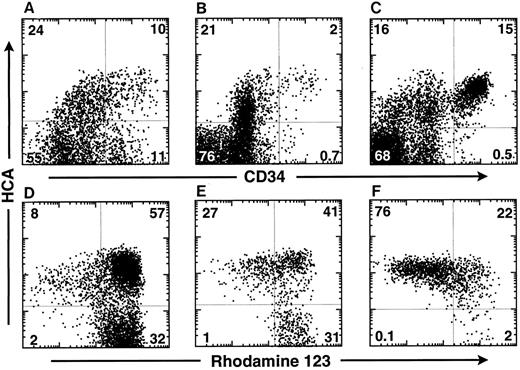
HCA is expressed at the surface of the most primitive human hematopoietic cells and maintained on their myeloid progeny.To determine whether HCA+ human hematopoietic cells include primitive progenitor/stem cells, FBM ABM, and MPB cells were triply-stained with the F84.1 and anti-CD34 MoAbs and with Rh123 dye. In the 18-week FBM, about 35% of cells expressed huHCA and CD34hi cells were subdivided into two distinct HCA+ and HCA− subsets (Fig 3A). Similarly, approximately 25% of ABM cells were HCA+, among which CD34+ HCA+ and CD34+ HCA− cell subsets were observed (Fig 3B). A similar percentage (32%) of HCA+ cells, including virtually all CD34 cells, was detected in one MPB sample (Fig 3C).
HCA expression by human hematopoietic cells in different tissues. Cells from 18-week FBM (A and D), ABM (B and E), and MPB from a multiple myeloma patient (C and F ) were stained as described in the Materials and Methods with Rh123 dye and MoAbs against CD34 and HCA. Isotype control staining was on all samples. The MPB sample was taken from day-2 leukapheresis, whereas the patient was treated with a high dose of cyclophosphamide and with GM-CSF. This particular MPB sample contained the highest percentage of CD34+ cells as compared with other leukapheresis harvests made at different days from the same patient. The percentage of cells in each quadrant is indicated. The bottom row (D, E, and F ) shows the Rh123 staining versus huHCA expression by CD34+ cells from each tissue analyzed above (A, B, and C).
HCA expression by human hematopoietic cells in different tissues. Cells from 18-week FBM (A and D), ABM (B and E), and MPB from a multiple myeloma patient (C and F ) were stained as described in the Materials and Methods with Rh123 dye and MoAbs against CD34 and HCA. Isotype control staining was on all samples. The MPB sample was taken from day-2 leukapheresis, whereas the patient was treated with a high dose of cyclophosphamide and with GM-CSF. This particular MPB sample contained the highest percentage of CD34+ cells as compared with other leukapheresis harvests made at different days from the same patient. The percentage of cells in each quadrant is indicated. The bottom row (D, E, and F ) shows the Rh123 staining versus huHCA expression by CD34+ cells from each tissue analyzed above (A, B, and C).
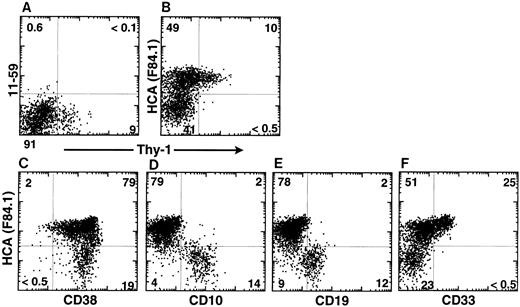
In the same experiment, CD34+ cells were further analyzed according to HCA expression and Rh123 staining. Rh123 can be used to separate distinct functional subsets of murine12,23 and human14,16,24,25 hematopoietic stem cells, of which the most primitive elements exhibit the lowest intracellular retention of that dye and are thus designated as Rh123lo. In the present analysis, a gate was then set to include only CD34+ cells with lymphoid-blast phenotype. In FBM, ABM, and MPB, virtually all Rh123lo-mid cells were observed to be HCA+; conversely, all HCA− cells were Rh123hi (Fig 3D, E, and F ). A large subset of HCA+ Rh123hi cells was present in each tissue. Therefore, all of the CD34+ cells characterized by a low retention of Rh123 express huHCA at their surface, which suggests that this antigen marks the most primitive human HSCs. This was investigated further by analyzing ABM CD34+ HCA+ cells for their coexpression of Thy-1, a marker of human early hematopoietic stem cells,1,14,25 and of CD38, a surface glycoprotein that is absent from the most immature (∼5%) ABM and FBM CD34+ cells but expressed by their differentiation-committed immediate progeny.1 26 All Thy-1–expressing CD34+ cells coexpressed huHCA (Fig 4B). Accordingly, most CD38−/lo cells were also huHCA+ (Fig 4C).
huHCA expression by subsets of CD34+ ABM cells. ABM was stained as described with MoAbs against CD34, Thy-1, rat KG-CAM (11-59; A), huHCA (F84.1; B), and isotype controls. FACS analyses of gated CD34+ populations are shown. MoAb 11-59 does not recognize human BM cells (A), whereas MoAb F84.1 binds a subset of CD34+ cells. Virtually all CD34+ Thy-1+ cells express huHCA (B). Alternatively, ABM cells were stained as described with MoAbs against CD34, huHCA, and, simultaneously, CD38 (C), CD10 (D), CD19 (E), and CD33 (F ). FACS analyses of gated CD34+ populations are shown. See text for details.
huHCA expression by subsets of CD34+ ABM cells. ABM was stained as described with MoAbs against CD34, Thy-1, rat KG-CAM (11-59; A), huHCA (F84.1; B), and isotype controls. FACS analyses of gated CD34+ populations are shown. MoAb 11-59 does not recognize human BM cells (A), whereas MoAb F84.1 binds a subset of CD34+ cells. Virtually all CD34+ Thy-1+ cells express huHCA (B). Alternatively, ABM cells were stained as described with MoAbs against CD34, huHCA, and, simultaneously, CD38 (C), CD10 (D), CD19 (E), and CD33 (F ). FACS analyses of gated CD34+ populations are shown. See text for details.
Yet, as can be expected from their high frequency, CD34+ HCA+ cells also include a majority of Rh123hi cells (Fig 3D, E, and F ), Thy-1− cells (Fig 4B) and CD38-bright cells (Fig 4C). These less immature HCA+ progenitors were further characterized by their expression of lineage-restricted differentiation antigens. As shown in Fig 4D and E, extremely few, if any, CD34+ HCA+ ABM cells coexpress the lymphoid-specific CD10 and CD19 markers,27 which are restricted to CD34+ HCA− cells. In contrast, about 33% of CD34+HCA+ cells were CD33+, and no cells positive for that myeloid lineage-restricted antigen were present among CD34+ HCA− cells (Fig 4F ).
We conclude that HCA is expressed at the surface of the most primitive human HSCs and maintained exclusively on their myeloid-committed immediate progeny.
In vitro functional characterization of the CD34+HCA+ cells subset from ABM.CD34+Lin− ABM cells were sorted by FACS first (Fig 5A) and were subdivided into four distinct populations on the basis of HCA expression and Rh123 retention (Fig 5B). They were cultured on SyS-1, a mouse BM stromal cell line permissive for the differentiation of primitive human HSCs into B-lymphoid and myeloid progenies.14,16 27 Rh123lo HCA+ (1 or 3 cells), Rh123mid HCA+ (1 or 3 cells), Rh123hi HCA+ (5 or 15 cells), and Rh123hi HCA− (5 or 15 cells) (Fig 5) were deposited into 96-well plates containing confluent SyS-1 cell monolayers cultured in the presence of LIF and IL-6, and the development of hematopoietic cobblestone areas was scored between 3 and 8 weeks of coculture. Strikingly, CAFC were totally absent from the HCA− Rh123hi population and present at a very low frequency (1/133) in HCA+ Rh123hi cells. CAFC frequency was significantly higher in HCA+ Rh123lo-mid populations. CAFC frequency in HCA+ Rh123mid cells peaked after 5 weeks of coculture and then declined to 1/24 at weeks 6 to 8. The highest CAFC frequency (1/11) was detected in HCA+ Rh123lo cells, although that subset was relatively quiescent at earlier scoring times (Fig 5C), indicating that HCA+ Rh123lo cells included the most primitive subpopulation. Indeed, after 6 weeks of coculture, multilineage populations, including CD34+, CD33+, and CD19+ cells, were present in some wells seeded with single HCA+ Rh123lo cells (Fig 5D and E).
In vitro proliferative potential of CD34+ Lin− cells further fractionated by huHCA expression and Rh123 staining. CD34+ Lin− ABM cells were sorted first (A). The sorted cells were stained with Rh123 as described and then for huHCA expression, based on isotope control. HCA+ Rh123lo (•), HCA+ Rh123mid (▴), HCA+ Rh123hi (▪), and HCA− Rh123hi (♦) (see sorting gates on [B]) were deposited into 96-well plates preseeded with SyS-1 stromal cells. CAFC growth was scored between 3 to 8 weeks of culture in two independent experiments (C). No cobblestone areas were detected in wells plated with HCA− Rh123hi cells. CAFC frequency (with lower 95% to upper 95% confidence interval) was determined by linear regression analysis as described in the Materials and Methods. CAFC frequency at 3 weeks in the HCA+ Rh123lo, HCA+ Rh123mid, and HCA+ Rh123hi cell subsets was 1/78 (50 to 174), 1/37 (26 to 67), and 1/266 (154 to 422), respectively. CAFC frequency at 4 weeks in the HCA+ Rh123lo, HCA+ Rh123mid, and HCA+ Rh123hi cell subsets was 1/18 (14 to 25), 1/16 (12 to 23), and 1/121 (89 to 188), respectively. CAFC frequency at 5 weeks in the HCA+ Rh123lo, HCA+ Rh123mid, and HCA+ Rh123hi cell subsets was 1/12 (9 to 15), 1/21 (16 to 31), and 1/185 (130 to 321), respectively. CAFC frequency at 6 to 8 weeks in the HCA+ Rh123lo, HCA+ Rh123mid, and HCA+ Rh123hi cell subsets was 1/11 (9 to 14), 1/24 (17 to 37), and 1/133 (98 to 208), respectively. On some occasions, proliferation of multilineage CD34+, CD33+, or CD19+ cell populations was detected; the example shown represents cells derived from HCA+ Rh123lo single cells deposited in culture 7 weeks earlier (D and E).
In vitro proliferative potential of CD34+ Lin− cells further fractionated by huHCA expression and Rh123 staining. CD34+ Lin− ABM cells were sorted first (A). The sorted cells were stained with Rh123 as described and then for huHCA expression, based on isotope control. HCA+ Rh123lo (•), HCA+ Rh123mid (▴), HCA+ Rh123hi (▪), and HCA− Rh123hi (♦) (see sorting gates on [B]) were deposited into 96-well plates preseeded with SyS-1 stromal cells. CAFC growth was scored between 3 to 8 weeks of culture in two independent experiments (C). No cobblestone areas were detected in wells plated with HCA− Rh123hi cells. CAFC frequency (with lower 95% to upper 95% confidence interval) was determined by linear regression analysis as described in the Materials and Methods. CAFC frequency at 3 weeks in the HCA+ Rh123lo, HCA+ Rh123mid, and HCA+ Rh123hi cell subsets was 1/78 (50 to 174), 1/37 (26 to 67), and 1/266 (154 to 422), respectively. CAFC frequency at 4 weeks in the HCA+ Rh123lo, HCA+ Rh123mid, and HCA+ Rh123hi cell subsets was 1/18 (14 to 25), 1/16 (12 to 23), and 1/121 (89 to 188), respectively. CAFC frequency at 5 weeks in the HCA+ Rh123lo, HCA+ Rh123mid, and HCA+ Rh123hi cell subsets was 1/12 (9 to 15), 1/21 (16 to 31), and 1/185 (130 to 321), respectively. CAFC frequency at 6 to 8 weeks in the HCA+ Rh123lo, HCA+ Rh123mid, and HCA+ Rh123hi cell subsets was 1/11 (9 to 14), 1/24 (17 to 37), and 1/133 (98 to 208), respectively. On some occasions, proliferation of multilineage CD34+, CD33+, or CD19+ cell populations was detected; the example shown represents cells derived from HCA+ Rh123lo single cells deposited in culture 7 weeks earlier (D and E).
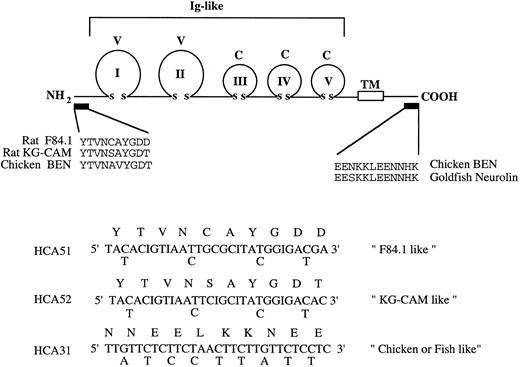
Molecular cloning of human HCA cDNA from CD34+ BM cells.At the outset of this work, the amino-terminal amino acid sequence but not the cDNA sequence of the rat F84.1 and KG-CAM antigens had been described and the cDNA sequences for likely homologues in chicken and fish (BEN/SC1/DM-GRASP/neurolin) were also known.8 18 An RT-PCR approach using a degenerate oligonucleotide (HCA51; Fig 6), based on the published N-terminal sequence of the F84.1 antigen as 5′-primer and oligo-dT as 3′-primer, was unsuccessful. Examination of the published N-terminal sequences of rat KG-CAM and F84.1 indicated two differences (Fig 6). Two degenerate oligonucleotide primers (HCA51 and HCA52) were therefore made according to their N-terminal sequences. Because no internal sequence was available for either F84.1 or KG-CAM, a 3′ primer (HCA31) was made based on the identical sequence of the cytoplasmic domains of chicken and fish BEN/SC1/DM-GRASP (Fig 6). Total cellular RNA was isolated from 105 CD34+ cells and oligo dT-primed cDNA was synthesized, from which HCA-related sequences were amplified with degenerate oligonucleotides by PCR. A major 1.4-kb cDNA fragment was identified in the PCR reaction containing the KG-CAM–like primers HCA52 and HCA31, but not in the reaction containing the F84.1-like primers HCA51 and HCA31 (data not shown). The 1.4-kb cDNA fragment was cloned into the pCRII vector and DNA sequencing showed that it encoded a partial open-reading frame with significant homology to the extracellular domain of BEN/SC1/DM-GRASP and so represented a likely candidate for HCA.
cDNA cloning strategy. The upper panel schematically illustrates the lg domain structure of chicken BEN and goldfish neurolin. The five extracellular V-type and C-type lg domains and transmembrane domain (TM) are shown. The amino-terminal sequences of rat F84.1 and KG-CAM (experimentally determined) and the deduced N-terminal sequence of chicken BEN are shown. The region of cytoplasmic sequence identity between chicken BEN and goldfish neurolin is shown at right. These amino acid sequences were used variously to derive the degenerate oligonucleotide primers HCA51, HCA52, and HCA31 shown below.
cDNA cloning strategy. The upper panel schematically illustrates the lg domain structure of chicken BEN and goldfish neurolin. The five extracellular V-type and C-type lg domains and transmembrane domain (TM) are shown. The amino-terminal sequences of rat F84.1 and KG-CAM (experimentally determined) and the deduced N-terminal sequence of chicken BEN are shown. The region of cytoplasmic sequence identity between chicken BEN and goldfish neurolin is shown at right. These amino acid sequences were used variously to derive the degenerate oligonucleotide primers HCA51, HCA52, and HCA31 shown below.
To clone the entire HCA coding region, the 1.4-kb cDNA clone was used to screen a cDNA library constructed from human CD34+ BM cells. Two cDNA clones, huHCA-1 and huHCA-2, approximately 2.1 kb and 2.3 kb in size, were derived and their DNA sequences determined for both strands. Confirmation of the 5′ sequence was also derived by 5′-PCR-RACE. Long open-reading frames encompassing 1752 nucleotides for huHCA-1 (Fig 7) and 1722 nucleotides for huHCA-2 were identified, which encoded polypeptides of 584 and 571 amino acid residues, respectively. huHCA-2 had an in-frame deletion of 13 amino acids between position 503 to 516 of huHCA-1, possibly resulting from alternative RNA splicing. huHCAs have the typical features of class I membrane proteins. Hydrophobic segments at the amino- and carboxy-terminal regions of the deduced sequences likely serve as signal peptide and transmembrane domain, respectively. In between, huHCA-1 and huHCA-2 contain five Ig-like domains (with typical intrachain disulfide bridges) and both huHCAs have a putative intracellular domain of 33 amino acids. The translated proteins have a calculated molecular weight of approximately 65 kD with 10 potential N-linked glycosylation sites in the extracellular region.
Deduced amino acid sequences of human HCA-1 and rat HCA aligned with chicken BEN/SC1/DM-GRASP. The single letter amino acid code is used. HCA-2 differed from HCA-1 by a deletion of 13 amino acids between positions 503-516 (▾). Stars denote conserved cysteine residues and conserved potential N-linked glycosylation sites are shaded. The predicted transmembrane domain is boxed. The positions of the oligonucleotides used for the cDNA cloning are shown (asterisks).
Deduced amino acid sequences of human HCA-1 and rat HCA aligned with chicken BEN/SC1/DM-GRASP. The single letter amino acid code is used. HCA-2 differed from HCA-1 by a deletion of 13 amino acids between positions 503-516 (▾). Stars denote conserved cysteine residues and conserved potential N-linked glycosylation sites are shaded. The predicted transmembrane domain is boxed. The positions of the oligonucleotides used for the cDNA cloning are shown (asterisks).
Alignment of huHCA-1 and huHCA-2 with chicken BEN/SC1/DM-GRASP showed that the proteins share 73% amino acid identity (Fig 7). Comparison of protein subsequences between huHCA and BEN/SC1/DM-GRASP indicated that the huHCA protein contains nearly all of the predicted structural features characteristic of BEN/SC1/DM-GRASP, including two V-type Ig domains, three C2-type Ig domains, identical sites for potential asparagine-linked glycosylation, and a particularly high degree of homology in the predicted transmembrane domain and cytoplasmic tail (95%). A 2.6-kb rat homologue of huHCA (raHCA) was obtained by screening a rat B49 cell cDNA library; its coding region shared 90% identity with huHCA and 72% identity with chicken BEN/SC1/DM-GRASP (Fig 7).
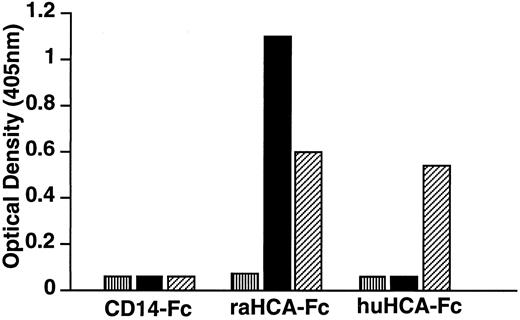
Recombinant HCA proteins are recognized by the MoAb F84.1.Fusion proteins were produced by directional cloning of the extracellular domain-encoding regions of huHCA or raHCA into the mammalian expression vector pCD5neg-1. The resulting constructs contain fused sequences encoding the CD5 signal sequence and the huHCA or raHCA extracellular domain-IgG1Fc fusion gene in tandem and direct the secretion of soluble huHCA-Fc or raHCA-Fc fusion proteins, respectively, which were purified from transfected COS-7 cell media. As tested by ELISA (Fig 8), MoAb F84.1 recognizes both purified huHCA-Fc and raHCA-Fc, whereas MoAb 11-59 recognizes raHCA-Fc but not huHCA-Fc. This result confirmed earlier observations that F84.1 recognized both rat and human hematopoietic cells, whereas 11-59 recognized only rat cells (Fig 2).
Immunochemical characterization of huHCA-Fc and raHCA-Fc fusion proteins. CD14-Fc, raHCA-Fc, and huHCA-Fc fusion proteins were purified as described and coated onto an ELISA plate at 10 μg/mL. Samples containing MoAbs (▪) 11-59, (▨) F84.1, or (▥) relevant controls (10 μg/mL IgG1) were added, followed by alkaline phosphatase conjugated with antimouse Ig for 1 hour. p-nitophenylphosphate was used to develop the ELISA and results were read at 405 nm using an ELISA reader.
Immunochemical characterization of huHCA-Fc and raHCA-Fc fusion proteins. CD14-Fc, raHCA-Fc, and huHCA-Fc fusion proteins were purified as described and coated onto an ELISA plate at 10 μg/mL. Samples containing MoAbs (▪) 11-59, (▨) F84.1, or (▥) relevant controls (10 μg/mL IgG1) were added, followed by alkaline phosphatase conjugated with antimouse Ig for 1 hour. p-nitophenylphosphate was used to develop the ELISA and results were read at 405 nm using an ELISA reader.
In different experiments, the 2.1-kb huHCA-1 and 2.6-kb raHCA cDNAs, containing the entire coding sequences, were cloned into the eukaryotic expression vector pSRa and the resulting constructs transfected into COS7 cells, which were labeled with 35S-methionine and -cysteine and analyzed by immunoprecipitation with MoAbs F84.1 and 11-59 (Fig 9). F84.1 recognized a 110-kD protein in both huHCA- and raHCA-transfected COS7 cells, whereas 11-59 recognized a 110-kD protein in the raHCA transfectants only. This result is consistent with the ELISA assay described above (Fig 8). The discrepancy between the predicted molecular weight of the unprocessed core protein (65 kD) and the apparent molecular weight detected by immunoprecipitation (∼100 kD) is presumed to be due to glycosylation. The neural cell F84.1 antigen is known to have different molecular weights in different cells, possibly as a result of varied levels of glycosylation.8
Immunoprecipitation of recombinant huHCA-1, huHCA-2, and raHCA transfected COS-7 cells with F84.1 and 11-59. COS-7 cells were transiently transfected with expression vectors encoding each HCA cDNA or with empty vector and immunoprecipitation and gel electrophoresis was performed as described in the Materials and Methods. A 3-day exposure of the dried gel is shown. The positions of molecular weight markers are shown at left.
Immunoprecipitation of recombinant huHCA-1, huHCA-2, and raHCA transfected COS-7 cells with F84.1 and 11-59. COS-7 cells were transiently transfected with expression vectors encoding each HCA cDNA or with empty vector and immunoprecipitation and gel electrophoresis was performed as described in the Materials and Methods. A 3-day exposure of the dried gel is shown. The positions of molecular weight markers are shown at left.
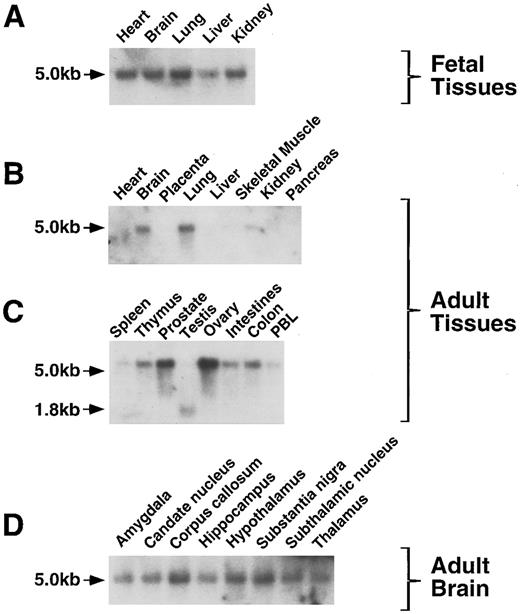
HCA expression in fetal and adult tissues.The expression of huHCA gene was analyzed in a variety of fetal and adult tissues by RNA (Northern) blotting. In human fetal tissues, huHCA mRNA was detected in heart, brain, lung, kidney, and, to a lesser extent, liver (Fig 10A). In adult tissues, huHCA is highly expressed in ovary and prostate, whereas expression in colon, intestine, thymus, spleen, and peripheral blood is relatively low (Fig 10B and C). A single message, approximately 5 kb in size, was identified in most of the fetal and adult tissues tested, with the exception of a 1.8-kb message found in adult testis. The expression of HCA was also detected in all the regions of adult brain that were analyzed (Fig 10D). HCA mRNA was not detected in adult heart and liver.
Analysis of HCA expression by RNA blot hybridization. RNA from a variety of fetal and adult human tissues was hybridized to an huHCA coding region probe. PBL, peripheral blood leukocytes. The positions of the 5.0- and 1.8-kb HCA transcripts are indicated.
Analysis of HCA expression by RNA blot hybridization. RNA from a variety of fetal and adult human tissues was hybridized to an huHCA coding region probe. PBL, peripheral blood leukocytes. The positions of the 5.0- and 1.8-kb HCA transcripts are indicated.
HCA mediates cell adhesion.To investigate whether the homophilic adhesion property of chicken BEN/SC1/DM-GRASP3 could be observed with huHCA, stable transfectants of CHO cells were obtained by cotransfection of pSRahuHCA-1 and pSV2-neo followed by G418 selection. HCA+ transfectants or parental cells were dissociated with EDTA and incubated either with or without agitation in EDTA/chondroitin sulphate/sodium azide. On incubation in still medium for 30 minutes, significant aggregation occurred in the HCA-transfected cell cultures (Fig 11, center panel) but not with control CHO cells (Fig 11, left panel). Furthermore, HCA-mediated adhesion could be inhibited with 40 μg/mL of recombinant soluble HCA-1-Fc (Fig 11, center panel), whereas the F84.1 antibody showed no inhibitory effect at 1 mg/mL (Fig 11, left panel). These results suggested that the epitope recognized by F84.1 was either not part of the adhesive binding site or the affinity of HCA for itself was much higher than for F84.1. Because HCA adhered in the presence of EDTA, divalent cations were not required, as was observed previously with chicken BEN/SC1/DM-GRASP.3
Adhesion assay of HCA transfectants. CHO cells transfected with huHCA-1 were incubated in the absence (center) or presence (right) of affinity-purified huHCA-1-Fc (40 μg/mL). Aggregation was observed by light microscopy after 30 minutes. Untransfected CHO cells did not aggregate in this condition (left).
Adhesion assay of HCA transfectants. CHO cells transfected with huHCA-1 were incubated in the absence (center) or presence (right) of affinity-purified huHCA-1-Fc (40 μg/mL). Aggregation was observed by light microscopy after 30 minutes. Untransfected CHO cells did not aggregate in this condition (left).
DISCUSSION
We have defined a novel surface marker of early human hematopoietic cells (huHCA), which is also expressed by subpopulations of developing neurons in the embryonic and early fetal human central nervous system. Homologues of HCA have previously been identified in chicken, fish, and rat tissues under the names of BEN/SC1/DM-GRASP, neurolin, and F84.1, respectively. Like its homologues, HCA is a member of the Ig superfamily that may mediate homophilic adhesion.
F84.1 antigen expression in the rat nervous system was similar to that of chicken BEN/SC1/DM-GRASP.2,6,8 Similarly, F84.1 staining of human embryo sections showed a distribution of HCA in the developing human central nervous system that is very close to that of BEN in the chicken. Amino-terminal sequencing of the rat F84.1 antigen showed similarity with the deduced amino-terminus of chicken BEN/SC1/DM-GRASP,8 of which rat KG-CAM was proposed to be another homologue.13 However, the expression of F84.1 and KG-CAM on human and rat hematopoietic cells (Fig 2) suggested that these antigens are different. To analyze the homology between HCA, F84.1, BEN/SC1/DM-GRASP, and KG-CAM, we cloned the HCA cDNA. Expression of raHCA and huHCA cDNAs showed conclusively that F84.1 and anti–KG-CAM recognize different epitopes on raHCA and that the KG-CAM epitope is not found on huHCA.
BEN/SC1/DM-GRASP is now known to be a cell surface antigen shared by cells of hematopoietic, neural, and a variety of other developing systems. Consistent with the various accounts of involvement of BEN/SC1/DM-GRASP in neuronal development is the high level of expression of HCA in human fetal and adult neural tissues observed by RNA blot hybridization (Fig 10). In the chicken, BEN/SC1/DM-GRASP is also expressed by immature CD4/CD8 thymocytes as well as by myeloid and erythroid progenitor cells.5 These observations suggested that BEN/SC1/DM-GRASP is a stage-specific rather than a lineage-specific marker expressed by immature hematopoietic cells. Activated avian T cells are also known to express high BEN levels.7 Ectopic expression of BEN/SC1/DM-GRASP induced by v-rel or c-rel is associated with avian B-cell lymphomas28; thus, ectopic expression of HCA might also contribute to human tumorigenesis.
However, whereas the bird embryo is an outstanding model in which to study many aspects of development, in vitro and in vivo assays for avian hematopoietic cells are very few in contrast with those available for mouse and human hematopoietic cells. Subsets of avian blood precursor cells are thus undefined, whereas a vast array of molecular markers are available for human blood cell lineages from early hematopoietic stem cells. We have taken advantage of those markers to document HCA expression along the hematopoietic hierarchy and detected this antigen at the surface of all the earliest forerunners of the human hematopoietic system, defined in ABM by CD34 and Thy-1 expression, absence or low levels of CD38, and low to medium Rh123 retention. Accordingly, the best performing CD34+ cells for cobblestone area formation on in vitro cultured BM stroma and for BM repopulation activity in SCID-hu mice (not shown) could be selected on the basis of HCA expression. The presence of HCA at the surface of the most primitive human HSCs can be paralleled with the observation that the ancestral hematopoietic progenitors that develop in paraaortic territories in the chicken embryo are BEN-positive.5
Yet, at variance with other molecules that mark mouse or human early blood stem cells, HCA remains expressed down the myeloid lineage-oriented hematopoietic hierarchy, whereas it disappears from the surface of lymphoid-committed CD34+ precursor cells coexpressing CD19. This observation could be extended to malignant hematopoiesis because myeloid leukemic blasts were generally found to be HCA+, whereas most lymphoid leukemias did not express HCA (F. Izhadifar-Legrand, unpublished observations). The role of HCA at the cell surface along that very pathway of blood cell differentiation is unknown. However, the documented role in homophilic adhesion of glycoproteins of the BEN family suggested a similar function for HCA. Indeed, we have further documented, but not yet proven, the hypothesis that HCA is a homophilic adhesion molecule by expression in CHO cells and competition with HCA-2-Fc (Fig 11). CD34+ cells coexpress many distinct adhesion molecules, some of which, like CD11a, CD11b, CD18, or CD62L, are differentially expressed on marrow-residing or circulating CD34+ cells (reviewed in Knapp et al1 ). HCA, too, was observed to be expressed by virtually all mobilized peripheral CD34+ cells tested, but by only a fraction of those in the fetal and adult BM. The expression of HCA by stromal cells in the fetal liver, thymus, and BM is under current investigation, the preliminary results of which indicate that this antigen is also present at the surface of subpopulations of nonhematopoietic cells that frame blood-forming tissues (F. Cortes, manuscript in preparation). Yet, preliminary studies have shown no effect of HCA-2-Fc on the outcome of in vitro assays of early hematopoietic development, including cobblestone-area formation by isolated HCA+ HSCs cultured on stromal cell lines (N.U. and Z.Y., unpublished observations). It remains to be determined if these results are due to insufficient amounts of HCA-Fc used, whether HCA can also bind other ligands of higher affinity than itself, or whether HCA does not serve as an important adhesion molecule in such long-term cultures. Similarly, HCA+ hematopoietic cells do not aggregate like the CHO-HCA transfectants. We speculate that HCA density on these cells is low and may require the presence of other factors (eg, activators of c-rel ) to induce high-level expression if HCA were to mediate homophilic adhesion of hematopoietic cells.
While this report was being prepared, a cDNA clone corresponding to the HCA sequence was published as ALCAM, an activated leukocyte-cell adhesion molecule.29 The investigators of that report also deduced that a ligand for ALCAM is CD6, a 130-kD type I membrane protein expressed by thymocytes and mature T cells. Thus, HCA may have at least two ligands, itself and CD6. It remains to be determined whether homophilic and heterophilic adhesion are both important for HCA+ hematopoietic progenitor function or if different ligands have different roles. We have tested the binding of anti-CD6 antibodies to hematopoietic progenitors and have observed low-level CD6 expression on CD34+ Rh123lo cells from ABM (not shown). Thus, HCA and CD6 are likely to be coexpressed on some cells. Future studies of these two ligands will be required to understand their role in the development of early hematopoietic cells in the BM and of T cells in the thymus.
ACKNOWLEDGMENT
We thank the following individuals for generously providing reagents: Dr B. Seed for the pCD5neg-1 plasmid and Dr E. Geisert Jr for the 11-59 antibody. We are grateful D. He, A. Friera, Kathy Wright, Steve Pronovost, and J. Schumann for expert technical assistance. We acknowledge Drs Stewart Craig, Benjamin Chen, Irving Weissman, Vladimir Bazil, and Bob Tushinski for sharing unpublished results, stimulating discussions, and critically reviewing the manuscript.
N.U. and Z.Y. contributed equally to this study.
Address reprint requests to Nobuko Uchida, PhD, SyStemix Inc, 3155 Porter Dr, Palo Alto, CA 94304.


![Fig. 5. In vitro proliferative potential of CD34+ Lin− cells further fractionated by huHCA expression and Rh123 staining. CD34+ Lin− ABM cells were sorted first (A). The sorted cells were stained with Rh123 as described and then for huHCA expression, based on isotope control. HCA+ Rh123lo (•), HCA+ Rh123mid (▴), HCA+ Rh123hi (▪), and HCA− Rh123hi (♦) (see sorting gates on [B]) were deposited into 96-well plates preseeded with SyS-1 stromal cells. CAFC growth was scored between 3 to 8 weeks of culture in two independent experiments (C). No cobblestone areas were detected in wells plated with HCA− Rh123hi cells. CAFC frequency (with lower 95% to upper 95% confidence interval) was determined by linear regression analysis as described in the Materials and Methods. CAFC frequency at 3 weeks in the HCA+ Rh123lo, HCA+ Rh123mid, and HCA+ Rh123hi cell subsets was 1/78 (50 to 174), 1/37 (26 to 67), and 1/266 (154 to 422), respectively. CAFC frequency at 4 weeks in the HCA+ Rh123lo, HCA+ Rh123mid, and HCA+ Rh123hi cell subsets was 1/18 (14 to 25), 1/16 (12 to 23), and 1/121 (89 to 188), respectively. CAFC frequency at 5 weeks in the HCA+ Rh123lo, HCA+ Rh123mid, and HCA+ Rh123hi cell subsets was 1/12 (9 to 15), 1/21 (16 to 31), and 1/185 (130 to 321), respectively. CAFC frequency at 6 to 8 weeks in the HCA+ Rh123lo, HCA+ Rh123mid, and HCA+ Rh123hi cell subsets was 1/11 (9 to 14), 1/24 (17 to 37), and 1/133 (98 to 208), respectively. On some occasions, proliferation of multilineage CD34+, CD33+, or CD19+ cell populations was detected; the example shown represents cells derived from HCA+ Rh123lo single cells deposited in culture 7 weeks earlier (D and E).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/8/10.1182_blood.v89.8.2706/4/m_bl_0047f5.jpeg?Expires=1763551959&Signature=fQBWMNA5szVAnUdsyYyyQsl~Ma2uXdeCccOZma6m25-PD9~4I3gbVBg3ImaRy2rWGj14hZApuW6xpKJATwf1jLW9IJZLZrYc2mmk088mI4BXhVPSs6c3HR65VzP-zpB1clHtNn3H5RIKte4bLIQE0hq4fMcb8b0yyT-PCosjYOXnBvyQIC0QMU06NDkLzrWgCcPEimWdwemWC1AayOTe0LdR~dVGZYScjDkdW0hffduc86busPKxhaI~9OIzGDOiRnek7-iUMGxY5SK3KSXzs-A4~hpTDJfy0DJr4cEjDHt3cS-MRU2kisLtsnBn7U6O2eM1Sh64ztG8mGKMNkMdqQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal