Abstract
Thirty-six patients with Philadelphia chromosome–positive acute lymphoblastic leukemia (Ph+ ALL) were studied for the presence of the bcr-abl fusion mRNA transcript after an allogeneic matched related (N = 12), partially matched related (N = 4), matched unrelated (N = 14), autologous (N = 5), or syngeneic (N = 1) bone marrow transplant (BMT). Seventeen were transplanted in relapse, and 19 were transplanted in remission. Twenty-three patients had at least one positive bcr-abl polymerase chain reaction (PCR) assay after BMT either before a relapse or without subsequent relapse. Ten of these 23 relapsed after a positive assay at a median time from first positive PCR assay of 94 days (range, 28 to 416 days). By comparison, only 2 relapses occurred in the 13 patients with no prior positive PCR assays; both patients had missed at least one scheduled follow-up assay and were not tested 2 months and 26 months before their relapse. The unadjusted relative risk (RR) of relapse associated with a positive PCR assay compared with a negative assay was 5.7 (95% confidence interval 1.2 to 26.0, P = .025). In addition, the data suggest that the type of bcr-abl chimeric mRNA detected posttransplant was associated with the risk of relapse: 7 of 10 patients expressing the p190 bcr-abl relapsed, compared with 1 of 8 who expressed only the p210 bcr-abl mRNA (P = .02, log-rank test). The RR of p190 bcr-abl positivity compared to PCR-negative patients was 11.2 (confidence interval 2.3-54.8, P = 0.003), whereas a positive test for p210 bcr-abl was apparently not associated with an increased relative risk. In separate multivariable models, PCR positivity remained a statistically significant risk factor for relapse after separately adjusting for donor (unrelated and partially matched v matched, autologous, and syngeneic), remission status at the time of transplant, the presence of acute graft-versus-host disease (GVHD), and type of conditioning regimen (total body irradiation dose of ≤1,200 cGy v <1,200 cGy). The PCR assay appears to be a useful test for predicting patients at high risk of relapse after BMT and may identify patients who might benefit from therapeutic interventions. The finding that the expression of p190 bcr-abl may portend an especially high risk of relapse suggests a different clinical and biologic behavior between p190 and p210 bcr-abl.
THE PHILADELPHIA chromosome (Ph) is the hallmark of chronic myeloid leukemia (CML) and also occurs in approximately 5% of children and 20% of adults with acute lymphoblastic leukemia (ALL).1-3 Ph-positivity in ALL is associated with aggressive disease and has been shown to be a poor prognostic factor, especially in children.1-3 Thus, Ph+ ALL patients are candidates for more aggressive treatment regimens such as bone marrow transplantation (BMT). Even with transplant the relapse rate for Ph+ ALL is quite high, ranging from 40% to 80%.4,5 Options for ALL patients who relapse after transplant are limited. Second transplant may be effective in children, but the prognosis is especially dismal for adults.6,7 Possible approaches to decrease relapse rates posttransplant such as adoptive immunotherapy, antibody-based therapies, or interferon8 9 exist, but all have their associated toxicities. Thus, it would be useful to be able to detect molecular evidence of recurrent disease before cytogenetic and hematologic relapse and offer intervention at that point.
Molecular analyses have established that the Ph translocation results in the joining of 3′ sequences of the tyrosine kinase c-ABL proto-oncogene on chromosome 9 to the 5′ sequences of the BCR gene on chromosome 22.10,11 In CML and some ALL cases the Ph breakpoint occurs within a 5.8-kb region on chromosome 22 known as the major breakpoint cluster region (M-bcr) of the BCR gene. This bcr-abl transcript encodes the chimeric p210 protein. However, in greater than 50% of Ph+ ALL cases the breakpoint in the BCR gene occurs 5′ to the M-bcr region, and the subsequent rearrangement with ABL yields a shorter 7.0-kb bcr-abl mRNA and expression of a 190-kD fusion protein.10-14 Both possible chimeric mRNAs (p210 bcr-abl and p190 bcr-abl) can be sensitively and specifically detected by the polymerase chain reaction (PCR).15 Recent studies have explored the utility of the PCR for detecting the bcr-abl fusion mRNA transcript in CML patients post-BMT,15-23 and it appears that the molecular detection of bcr-abl often predicts subsequent relapse. Several centers have thus embarked on clinical trials using the molecular detection of bcr-abl as a trigger to initiate intervention with agents such as donor lymphocytes or interferon. Although there is a large body of literature documenting the efficacy of bcr-abl detection in CML, little data exist on the detection of minimal residual disease (MRD) in Ph+ ALL post-BMT.24-26 In this study we explored the clinical utility of the PCR detection of bcr-abl post-BMT and examined if the risk varied with the specific molecular breakpoint (p190 v p210 bcr-abl).
MATERIALS AND METHODS
Patients.All patients with ALL referred to the Fred Hutchinson Cancer Research Center for BMT were eligible for study. Marrow samples were intended to be collected pretransplant and then at days 21, 56, and 80 post-BMT, and at 6-month intervals thereafter.
PCR amplification of BCR-ABL transcripts.RNA isolation from the cryopreserved specimens and the conditions and specific primer sequences used in reverse transcription/PCR amplification were exactly as previously described.15 27 Specimens were analyzed using three independent sets of amplification primers. One set of “nested” primers was designed to detect rearrangements within the major breakpoint region of the BCR gene (M-bcr). The chimeric mRNA generated by this rearrangement is referred to here as p210 bcr-abl. The expected PCR product size after amplification with these primers are 304 or 234 bp, depending on the presence or absence of BCR exon b3. Another set of nested primers was designed to be complementary to BCR exon I and ABL exon II. This PCR assay yielded a 190-bp PCR product that is designated as p190 bcr-abl. An aliquot of each RNA sample was also reverse transcribed (RT) and PCR amplified using a set of primers designed to amplify sequences from the ubiquitously expressed β2-microglobulin gene. Positive amplification of this control gene tested the possibility of a false-negative bcr-abl RT/PCR assay resulting from an inadequate specimen, poor RNA isolation, or failed reverse transcription or PCR amplification. Identification of bcr-abl fusion transcripts was based on the size of PCR products.
All bcr-abl RT/PCR analyses were performed without knowledge of the patient's clinical data and cytogenetic status. All specimens were scored for positivity for the p210 bcr-abl and p190 bcr-abl fusion transcript with a β2-microglobulin PCR amplification to test for the integrity of the RNA. Positive controls were the ALL-1 cell line for the p190 bcr-abl and the K562 cell line for p210 bcr-abl. The sensitivities of the p190 and p210 nested PCR assays detect at least a 10−5 dilution of the cell line mRNA in a background of normal mRNA. Thus, the PCR assays 10−5 dilutions of ALL-1 and K562 were used as positive controls, and any if any set of reactions did not reach this level of sensitivity, the entire set of assays were repeated. Any sample that did not have a positive β2-microglobulin PCR reaction was declared uninterpretable and excluded from subsequent statistical analyses.
The PCR protocol included a number of specific procedures to decrease the possibility of PCR product carryover and contamination.28 These included separate rooms for nucleic acid isolation, PCR set-up, PCR amplification, and Southern blotting; the use of dedicated pipettes and filtered pipette tips for all steps of the PCR assay; and the inclusion of a nonnucleic acid containing “blank” during all PCR reactions to control for reagent contamination.
Cytogenetic analysis and fluorescence in situ hybridization. Chromosomes were prepared from direct and 24- to 48-hour unstimulated BM culture using standard techniques and analyzed following trypsin Giemsa banding. Y chromosome–specific in situ hybridization was performed on samples from patients following sex mismatched BMT. Standard cytogenetic preparations of BM were fixed onto clean glass slides, denatured in 70% formamide, 2× SSC at 70°C for 3 minutes (1× SSC = 0.15 mol/L NaCl/0.015 mol/L sodium citrate), dehydrated through ethanol, and air dried. Biotin-labeled plasmid pY3.4 containing Y chromosome–specific sequences (a gift from Dr Y.-F. Lau, University of California, San Francisco) were denatured by incubation at 70°C for 5 minutes, applied to the slides under a coverslip, and allowed to hybridize overnight at 37°C. Unhybridized probe was removed by washes in 50% formamide, 1× SSC at 37°C for 30 minutes then in 2× SSC and 1× SSC, each for 30 minutes at room temperature. Hybridized probe was detected with Fluorescein Avidin (Vector Laboratories, Burlingame, CA) and nuclei were counterstained with propidium iodide (Sigma, St Louis, MO) in antifade reagent DABCO (Sigma). Analysis of at least 200 interphase nuclei for Y chromatin signal was performed using a Zeiss Axioscope epifluorescence microscope (Carl Zeiss, Inc, Thornwood, NY) and percentage of donor versus host cells was calculated.
Direct nucleotide sequencing of PCR products.The identity of p190 and p210 bcr-abl PCR products was in some circumstances verified by direct nucleotide sequencing using the Applied Biosystems Inc (ABI; Foster City, CA) fluorescence sequencing system using labeled dideoxynucleotides. The sequencing reaction was performed in a Perkin Elmer Cetus (Foster City, CA) thermocycler with cycling parameters of 94°C × 1 minute, 50°C × 30 seconds, and 60°C × 2 minutes for a total of 25 cycles using the PCR primers as sequencing primers. For these reactions approximately 40 ng of PCR product was used with the addition of 10 ng of primer. The sequencing reaction products where then purified using a Centri-Sep column (Princeton Separation, Adelphia, NJ) and the products loaded on a sequencing gel and analyzed with the ABI florescent sequencer.
Transplant regimen.Thirty-two patients received a conditioning regimen of cyclophosphamide 120 mg/kg over 2 successive days with total body irradiation of 1,200 to 1,575 cGY from dual opposing cobalt sources (5 patients received ≤1,200 cGy, and 27 received >1,200 cGy). Four patients received a preparative regimen of carmustine, cyclophosphamide, and etoposide. The two patients receiving mismatched related transplants also received antithymocyte globulin (ATG) during their preparative regimen. All patients received graft-versus-host disease (GVHD) prophylaxis of methotrexate and cyclosporine as previously described.29
Definitions of outcomes and statistics.The endpoint of relapse was defined as either the presence of leukemic blasts in the BM or peripheral blood or evidence of the Ph chromosome on cytogenetic examination. A positive PCR assay was defined as a positive signal of the appropriate size in the posttransplant sample with simultaneous positive signal in the original diagnostic marrow and having no signal in both normal BM and non-nucleic acid containing PCR mix. In the event that patients had intermittent PCR-positivity the patient was declared as PCR-positive at the time of first PCR-positive assay.
Relapse rates for patients classified by PCR test result at the stated time intervals were estimated by the cumulative incidence statistic.30 Patients whose PCR results were positive concurrently with a sample which showed hematologic relapse were classified according to the PCR status at the last test before relapse. Thus, patients no. 6350 and 6334, who had a negative PCR assay before subsequently relapsing with a concurrent positive PCR assay (on days 71 and 856, respectively), were considered as relapses in the PCR-negative group.
To compare the hazard rates of relapse, PCR status was modeled as a time-dependent covariate in a Cox regression model.30 Thus, a patient was classified as PCR-negative until the occurrence of a PCR-positive test, at which point the patient was subsequently classified as positive. The data were also considered to be left-truncated; ie, a patient did not enter the risk set until the first PCR test was performed. This allowed the variable entry times onto study to be accommodated. All P values are two-sided and those in the regression models were derived from the Wald test; no adjustments were made for multiple comparisons.31 The grading of acute and chronic GVHD has been described.29
RESULTS
Patients.Thirty-six patients had evidence of Ph+ ALL before transplant. Seventeen patients had a BMT during relapse, while 19 patients were transplanted in remission. Twelve patients had an allogeneic matched donor, 4 a mismatched donor, 14 received unrelated donor transplants, 5 patients had an autologous transplant, and 1 a syngeneic transplant. The median age of the 36 patients was 31 years (range, 1 to 56 years). The patient characteristics and clinical course are summarized in Table 1.
Patient Characteristics
| UPN . | Age/Sex . | Donor . | Phase . | bcr-abl . | aGVHD . | cGVHD . | Relapse . | DFS . |
|---|---|---|---|---|---|---|---|---|
| 5114 | 40/M | Auto | Rel. | p190 | 0 | − | d199 | — |
| 5191 | 24/M | URD | Rem. | — | 3 | + | — | >2,194 |
| 5291 | 3/F | Rel-mm | Rel. | p190 | 2 | + | — | >2,205 |
| 5305 | 31/F | Rel= | Rem. | p190 | 0 | − | d126 | — |
| 5476 | 41/F | Rel= | Rem. | p210 | 0 | − | — | >2,139 |
| 5509 | 43/F | Rel= | Rem. | p210 | 1 | − | d113 | — |
| 5579 | 15/F | Rel= | Rel. | — | 2 | + | — | (X564) |
| 5601 | 9/F | Auto | Rem. | — | 0 | − | — | >1,854 |
| 5774 | 4/F | Rel-mm | Rem. | p190 | 3 | + | — | >1,847 |
| 5942 | 35/F | URD | Rel. | p190/210 | 4 | NE | — | (X74) |
| 6007 | 29/F | Rel= | Rem. | — | 2 | + | — | >1,766 |
| 6056 | 18/F | URD | Rel. | p190/210 | 3 | NE | d96 | — |
| 6112 | 19/M | Rel= | Rel. | p190/210 | 3 | + | — | >1,475 |
| 6186 | 19/M | Auto | Rem. | p190/210 | 0 | − | d443 | — |
| 6323 | 49/M | Rel= | Rel. | p190/210 | 1 | NE | — | (X71) |
| 6334 | 55/M | Auto | Rem. | — | 0 | − | d860 | — |
| 6350 | 40/F | Twin | Rel. | — | 2 | − | d71 | — |
| 6380 | 6/M | Rel= | Rel. | — | 2 | − | — | >1,484 |
| 6641 | 38/F | Rel= | Rem. | — | 0 | + | — | (X472) |
| 6642 | 7/F | URD | Rel. | p190 | 4 | NE | — | (X60) |
| 6968 | 40/M | Rel= | Rem. | p190 | 2 | − | d134 | — |
| 7055 | 15/M | Auto | Rem. | — | 0 | − | — | >1,201 |
| 7361 | 47/F | URD | Rem. | p210 | 2 | + | — | >1,120 |
| 7384 | 26/M | Rel= | Rel. | p190 | 2 | NE | d46 | — |
| 7944 | 34/M | Rel-mm | Rel. | p210 | 2 | NE | — | (X94) |
| 7958 | 20/M | URD | Rem. | p210 | 2 | + | — | >820 |
| 8124 | 26/M | URD | Rem. | p210 | 2 | − | — | >601 |
| 8765 | 47/F | URD | Rel. | p190 | 2 | − | d144 | — |
| 9115 | 3/F | URD | Rem. | — | 3 | + | — | >423 |
| 9222 | 1/M | URD | Rem. | — | 2 | + | — | >405 |
| 9362 | 37/M | Rel-mm | Rel. | p210 | 2 | + | — | >418 |
| 9552 | 35/M | Rel= | Rel. | p210 | 2 | − | d141 | — |
| 9635 | 29/M | URD | Rel. | p210 | 2 | + | — | >267 |
| 9833 | 34/M | URD | Rem. | — | 2 | + | — | >134 |
| 9861 | 51/M | URD | Rem. | — | 2 | − | — | >173 |
| 10029 | 39/F | URD | Rel. | p190 | 2 | NE | d74 | — |
| UPN . | Age/Sex . | Donor . | Phase . | bcr-abl . | aGVHD . | cGVHD . | Relapse . | DFS . |
|---|---|---|---|---|---|---|---|---|
| 5114 | 40/M | Auto | Rel. | p190 | 0 | − | d199 | — |
| 5191 | 24/M | URD | Rem. | — | 3 | + | — | >2,194 |
| 5291 | 3/F | Rel-mm | Rel. | p190 | 2 | + | — | >2,205 |
| 5305 | 31/F | Rel= | Rem. | p190 | 0 | − | d126 | — |
| 5476 | 41/F | Rel= | Rem. | p210 | 0 | − | — | >2,139 |
| 5509 | 43/F | Rel= | Rem. | p210 | 1 | − | d113 | — |
| 5579 | 15/F | Rel= | Rel. | — | 2 | + | — | (X564) |
| 5601 | 9/F | Auto | Rem. | — | 0 | − | — | >1,854 |
| 5774 | 4/F | Rel-mm | Rem. | p190 | 3 | + | — | >1,847 |
| 5942 | 35/F | URD | Rel. | p190/210 | 4 | NE | — | (X74) |
| 6007 | 29/F | Rel= | Rem. | — | 2 | + | — | >1,766 |
| 6056 | 18/F | URD | Rel. | p190/210 | 3 | NE | d96 | — |
| 6112 | 19/M | Rel= | Rel. | p190/210 | 3 | + | — | >1,475 |
| 6186 | 19/M | Auto | Rem. | p190/210 | 0 | − | d443 | — |
| 6323 | 49/M | Rel= | Rel. | p190/210 | 1 | NE | — | (X71) |
| 6334 | 55/M | Auto | Rem. | — | 0 | − | d860 | — |
| 6350 | 40/F | Twin | Rel. | — | 2 | − | d71 | — |
| 6380 | 6/M | Rel= | Rel. | — | 2 | − | — | >1,484 |
| 6641 | 38/F | Rel= | Rem. | — | 0 | + | — | (X472) |
| 6642 | 7/F | URD | Rel. | p190 | 4 | NE | — | (X60) |
| 6968 | 40/M | Rel= | Rem. | p190 | 2 | − | d134 | — |
| 7055 | 15/M | Auto | Rem. | — | 0 | − | — | >1,201 |
| 7361 | 47/F | URD | Rem. | p210 | 2 | + | — | >1,120 |
| 7384 | 26/M | Rel= | Rel. | p190 | 2 | NE | d46 | — |
| 7944 | 34/M | Rel-mm | Rel. | p210 | 2 | NE | — | (X94) |
| 7958 | 20/M | URD | Rem. | p210 | 2 | + | — | >820 |
| 8124 | 26/M | URD | Rem. | p210 | 2 | − | — | >601 |
| 8765 | 47/F | URD | Rel. | p190 | 2 | − | d144 | — |
| 9115 | 3/F | URD | Rem. | — | 3 | + | — | >423 |
| 9222 | 1/M | URD | Rem. | — | 2 | + | — | >405 |
| 9362 | 37/M | Rel-mm | Rel. | p210 | 2 | + | — | >418 |
| 9552 | 35/M | Rel= | Rel. | p210 | 2 | − | d141 | — |
| 9635 | 29/M | URD | Rel. | p210 | 2 | + | — | >267 |
| 9833 | 34/M | URD | Rem. | — | 2 | + | — | >134 |
| 9861 | 51/M | URD | Rem. | — | 2 | − | — | >173 |
| 10029 | 39/F | URD | Rel. | p190 | 2 | NE | d74 | — |
In the Donor column, “Auto,” “Rel=,” “Rel-mm,” and “URD” signify an autologous, matched-related, mismatched-related, and unrelated donor, respectively. In the Phase category, “Rem.” and “Rel.” refer to the patient being in remission or relapse at the time of the transplant. “p190” and “p210” denote the type of bcr-abl transcript found posttransplant. In the aGVHD column the grading of acute GVHD is based from 0-4. A “+” in the cGVHD category denotes clinically extensive chronic GVHD, while NE designates patients not evaluable. In the Relapse category the day of relapse post-BMT is given, if appropriate. In the DFS column, “>” denotes the last day of contact with the patient disease-free. The “X” denotes a nonleukemic death and the date of that death.
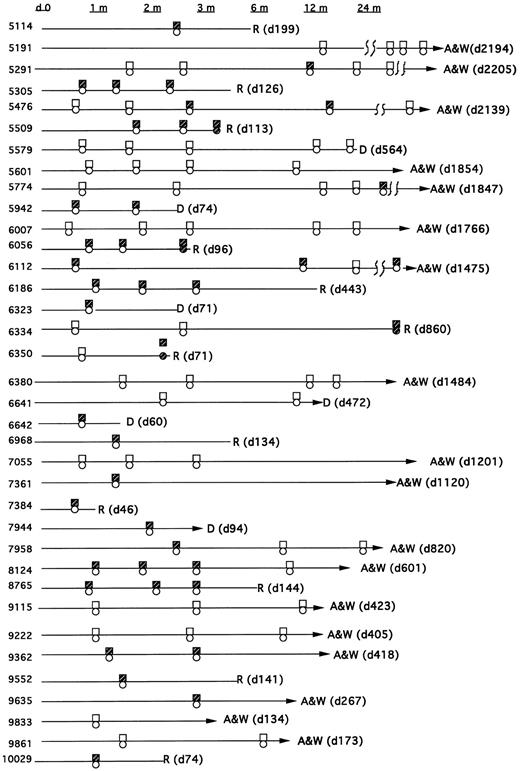
Types of bcr/abl fusion transcripts.All patients were monitored for both the p210 bcr-abl and p190 bcr-abl fusion transcript. In sum, 23 patients had at least one positive PCR assay after their transplant and before relapse (or without relapse), whereas 13 had persistently negative assays (Fig 1). Eight patients had only the p210 transcript post-BMT, and 10 had only the p190 bcr-abl transcript. Five patients showed evidence of both transcripts post-BMT (though not necessarily simultaneously). In all five of these patients the PCR assays were repeated to confirm these results and to exclude the possibility that amplimer contamination could have caused a false-positive test. In addition, both p190 and p210 bcr-abl PCR products were sequenced in these patients to confirm their identity.
Time course of patients post-BMT showing PCR, cytogenetics, and pathology results for detection of bcr-abl, the Philadelphia chromosome, or morphological relapse. The boxes reflect the PCR assay (open = −; closed = +). The circles reflect cytogenetic or hematologic relapse (open = remission; closed = relapse).
Time course of patients post-BMT showing PCR, cytogenetics, and pathology results for detection of bcr-abl, the Philadelphia chromosome, or morphological relapse. The boxes reflect the PCR assay (open = −; closed = +). The circles reflect cytogenetic or hematologic relapse (open = remission; closed = relapse).
Thirteen patients were persistently PCR-negative. Five of these patients were transplanted in remission but were PCR-positive before BMT. Eight of these patients were either PCR-negative before transplant (n = 4) or were not tested (n = 4). However, all eight had diagnostic cytogenetics at their referral institutions which showed the Ph chromosome.
MRD detection and outcome.Of the 13 patients who were persistently PCR-negative, 8 are alive and well, 3 died of nonleukemic causes at days 31, 472, and 564 post-BMT, and 2 have relapsed. In these 2 relapsing patients (unique patient number [UPN] 6350 and 6334), the first positive PCR occurred simultaneous with the discovery of relapse, and thus for these analyses they are classified as relapses in the PCR-negative group. In patient 6350, a PCR assay was negative on day 22, the day 56 marrow was not collected, and the later routine marrow on day 71 revealed PCR-positivity and pathologic relapse. In patient 6334 all routine marrows were negative, the last on day 76. The patient was then not studied until day 856, at which time a PCR assay, as well as gross pathology, revealed relapse. Of the 23 patients with a positive PCR test before relapse (or without subsequent relapse), 10 relapsed and 2 died of nonleukemic causes.
Considering PCR status as a time-dependent covariate, the relative risk of relapse for PCR-positive patients compared with PCR-negative patients was 5.7 (95% confidence interval [CI], 1.2 to 26.0, P = .025). The median time from first PCR-positive test to relapse was 94 days (range, 28 to 416 days).
The analysis was further performed by the type of bcr-abl (p190 v p210) expressed anytime post-BMT. Of the 10 patients with only p190 bcr-abl mRNA detected post-BMT, 7 relapsed, 1 died of nonleukemic causes on day 60, and 2 patients (5774 and 5291) are disease-free at 1,847 and 2,205 days. The relative risk (RR) of relapse of p190 bcr-abl detection compared to PCR-negativity was 11.2 (CI 2.3 to 54.8, P = .003). In the 8 patients who had only p210 bcr-abl mRNA post-BMT, 1 relapsed, 1 died of nonleukemic causes (day 94), and 6 are alive and disease-free from days 267-2,139. The RR of relapse for p210 bcr-abl compared with PCR-negativity was 1.0 (CI 0.1 to 10.9). In these 6 disease-free survivors the median time from first p210 bcr-abl PCR-positivity to last follow-up is 568 days. Lastly, 5 patients showed both p190 bcr-abl and p210 bcr-abl, and 2 relapsed, 2 died of nonleukemic causes (days 71 and 74), and 1 (6112) is alive and disease-free 1475 days after transplant.
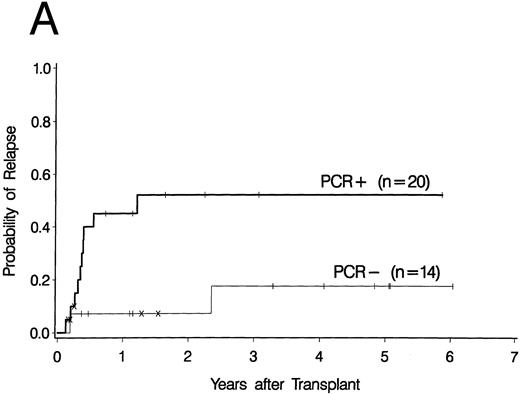
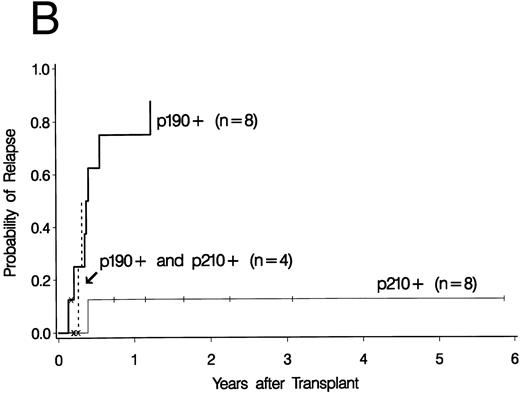
Cumulative incidence curves for relapse were constructed for the 34 patients who had a PCR test before 100 days post-BMT. At 3 years post-BMT the relapse rate for patients having a PCR-positive test before day 100 was 52% compared with 17% for patients having all PCR-negative tests before day 100 (Fig 2A). A breakdown of the positive patient's type of bcr-abl detected reveals an 88% relapse rate for p190 bcr-abl, compared with 12% for p210 bcr-abl positive patients and 50% for patients expressing both p190 and p210 bcr-abl (Fig 2B).
Upper curves (A) show the cumulative incidence of relapse by the presence or absence of bcr-abl detected by PCR before day 100 post-BMT. Lower curves (B) show the cumulative incidence of relapse by the type of bcr-abl transcript found after transplant. Ticks indicate patients alive without relapse and “×” marks denote patients who died without relapse.
Upper curves (A) show the cumulative incidence of relapse by the presence or absence of bcr-abl detected by PCR before day 100 post-BMT. Lower curves (B) show the cumulative incidence of relapse by the type of bcr-abl transcript found after transplant. Ticks indicate patients alive without relapse and “×” marks denote patients who died without relapse.
Clinical features that might have influenced the relapse rate were analyzed by both univariable and multivariable models. In the univariable models PCR-positivity and conditioning regimen were associated with the hazard of relapse (Table 2). The effect of chronic GVHD (cGVHD) was difficult to assess statistically. Fourteen patients developed cGVHD, and none relapsed. However, of these 14, 7 had positive PCR assays, while 7 had persistently negative assays for bcr-abl. For multivariable analyses we included one of several variables to PCR status, thereby generating separate models, one for each of these variables in addition to the PCR status (Table 3). Thus, the remission status (remission versus relapse at the time of BMT), the type of transplant (unrelated and partially matched v matched, autologous, or syngeneic), the presence of grades ≥II acute GVHD, and the conditioning regimen (chemotherapy only or total body irradiation [TBI]-containing with ≤1,200 cGy v TBI-containing with >1,200 cGy) were added separately to multivariable models containing the PCR status. Because of the limited number of events (relapses), the multivariable models were restricted to just two variables. A positive PCR assay remained statistically significantly associated with relapse after adjustment for these variables.
Univariable Analysis of Relapse
| Variable . | Relative Risk . | 95% CI . | P Value . |
|---|---|---|---|
| PCR status | |||
| Negative | 1.0 | ||
| p190 or p210+ | 5.7 | 1.2-26.0 | .025 |
| PCR status | |||
| Negative | 1.0 | ||
| p190+ | 11.2 | 2.3-54.8 | .003 |
| Donor type | |||
| Auto, twin, Rel= | 1.0 | ||
| URD, Rel-mm | 0.4 | 0.1-1.4 | .133 |
| Acute GVHD | |||
| Grades 0-1 | 1.0 | ||
| Grades 2-4 | 0.6 | 0.2-2.0 | .427 |
| Phase of disease | |||
| Remission | 1.0 | ||
| Relapse | 2.6 | 0.8-8.3 | .106 |
| Conditioning | |||
| TBI > 1,200 cGY | 1.0 | ||
| Chemo only or <1,200 cGy | 4.6 | 1.5-14.5 | .008 |
| Variable . | Relative Risk . | 95% CI . | P Value . |
|---|---|---|---|
| PCR status | |||
| Negative | 1.0 | ||
| p190 or p210+ | 5.7 | 1.2-26.0 | .025 |
| PCR status | |||
| Negative | 1.0 | ||
| p190+ | 11.2 | 2.3-54.8 | .003 |
| Donor type | |||
| Auto, twin, Rel= | 1.0 | ||
| URD, Rel-mm | 0.4 | 0.1-1.4 | .133 |
| Acute GVHD | |||
| Grades 0-1 | 1.0 | ||
| Grades 2-4 | 0.6 | 0.2-2.0 | .427 |
| Phase of disease | |||
| Remission | 1.0 | ||
| Relapse | 2.6 | 0.8-8.3 | .106 |
| Conditioning | |||
| TBI > 1,200 cGY | 1.0 | ||
| Chemo only or <1,200 cGy | 4.6 | 1.5-14.5 | .008 |
Univariable analysis of the relative risk of relapse associated with PCR status, donor type, acute and chronic GVHD, remission status at the time of transplant, and type of conditioning regimen. In the chronic GVHD category, “positive” means clinically extensive disease.
Multivariable Analysis of Risk Factors for Relapse
| . | Risk Factor . | RR . | 95% CI . | P Value . |
|---|---|---|---|---|
| Model 1 | PCR Status | |||
| PCR-negative | 1.0 | — | — | |
| PCR-positive | 4.8 | 1.0-22.7 | .046 | |
| Remission Status | ||||
| Remission | 1.0 | — | — | |
| Relapse | 2.0 | 0.6-6.6 | .249 | |
| Model 2 | PCR Status | |||
| PCR-negative | 1.0 | — | — | |
| PCR-positive | 6.9 | 1.5-32.9 | .014 | |
| Donor Type | ||||
| Matched-related | 1.0 | — | — | |
| URD, mismatch | 0.3 | 0.1-1.0 | .058 | |
| Model 3 | PCR Status | |||
| PCR-negative | 1.0 | — | — | |
| PCR-positive | 5.8 | 1.3-26.6 | .023 | |
| GVHD | ||||
| Grades 0-I (or Auto) | 1.0 | — | — | |
| Grades ≥ II | 0.6 | 0.2-2.0 | .410 | |
| Model 4 | PCR Status | |||
| PCR-negative | 1.0 | — | — | |
| PCR-positive | 5.5 | 1.2-25.4 | .028 | |
| TBI | ||||
| >1200 cGy | 1.0 | — | — | |
| Chemo or ≤1,200 | 4.3 | 1.3-13.8 | .015 |
| . | Risk Factor . | RR . | 95% CI . | P Value . |
|---|---|---|---|---|
| Model 1 | PCR Status | |||
| PCR-negative | 1.0 | — | — | |
| PCR-positive | 4.8 | 1.0-22.7 | .046 | |
| Remission Status | ||||
| Remission | 1.0 | — | — | |
| Relapse | 2.0 | 0.6-6.6 | .249 | |
| Model 2 | PCR Status | |||
| PCR-negative | 1.0 | — | — | |
| PCR-positive | 6.9 | 1.5-32.9 | .014 | |
| Donor Type | ||||
| Matched-related | 1.0 | — | — | |
| URD, mismatch | 0.3 | 0.1-1.0 | .058 | |
| Model 3 | PCR Status | |||
| PCR-negative | 1.0 | — | — | |
| PCR-positive | 5.8 | 1.3-26.6 | .023 | |
| GVHD | ||||
| Grades 0-I (or Auto) | 1.0 | — | — | |
| Grades ≥ II | 0.6 | 0.2-2.0 | .410 | |
| Model 4 | PCR Status | |||
| PCR-negative | 1.0 | — | — | |
| PCR-positive | 5.5 | 1.2-25.4 | .028 | |
| TBI | ||||
| >1200 cGy | 1.0 | — | — | |
| Chemo or ≤1,200 | 4.3 | 1.3-13.8 | .015 |
Four multivariable models examining the effect of the PCR test for bcr-abl while controlling for other clinical variables. In each model the relative risk (RR) of relapse compares PCR-positive to PCR-negative patients. In addition, clinical variables are added separately to the model containing the PCR status of the patient. For the variable of remission status at the time of BMT, relapse is compared to remission; for donor type, unrelated or partially matched v matched, syngeneic, and autologous; for GVHD, grades II-IV acute GVHD v grades O-I; and for conditioning regimen, patients receiving chemotherapy only or TBI-containing regimens with ≤1,200 cGy v TBI-containing regimens with >1,200 cGy.
The correlation of BM and peripheral blood results.We received simultaneous BM and peripheral blood samples on 31 occasions. The PCR results were concordant 23 times (74%), while in 5 (16%) cases the BM was positive while the peripheral blood was negative. In 3 cases (10%) the peripheral blood tests were positive with negative BM tests.
DISCUSSION
In this study, we found that patients with the presence of the bcr-abl chimeric mRNA after marrow transplant for Ph+ ALL had a high risk of subsequent relapse compared to patients without detectable bcr-abl (RR = 5.7, P = .025). The prognostic importance of the PCR assay remained after controlling for other clinical variables that might influence relapse risk. The median time from first PCR-positivity to relapse was 94 days, a large enough “window” of time in which to launch therapeutic intervention. The strongest association between bcr-abl expression and relapse was in the patients who expressed the p190 bcr-abl post-BMT. Of the 12 relapses (10 in PCR-positive and 2 in PCR-negative patients), only 1 occurred in a patient who expressed p210 bcr-abl alone. Expression of both p190 and p210 bcr-abl, recently found to occur in approximately 10% of Ph+ ALL,32 was seen in 5 of our patients, and 2 of these patients relapsed, again suggesting that the presence of p190 bcr-abl confers a high risk of relapse.
The results also suggest that the biology of p190 and p210 bcr-abl leukemia might differ. Studies have shown that the p190 BCR-ABL chimeric protein compared with the p210 protein has greater in vitro tyrosine kinase activity,33 and produces a more virulent pattern of leukemia in both the mouse transplantation model and transgenic mice systems.34,35 In addition, clinical data show that the Philadelphia chromosome may convey a worse prognosis in children than in adults.27 36 Children with Ph+ ALL have predominately p190 bcr-abl expression whereas adults with ALL express p190 or p210 bcr-abl nearly equally. This difference may account for the poorer prognosis for children with Ph+ ALL because of the biologic behavior of p190 bcr-abl. Moreover, we have previously published data that suggest that Ph+ adult patients bearing the p210 bcr-abl at diagnosis may have a different response to conventional chemotherapy than patients bearing the p190 bcr-abl.27 Complete responses (CR) occurred in only 5 of 14 patients who expressed p210 bcr-abl, but the median relapse-free survival of these patients achieving a CR has yet to be reached. In contrast, 6 of 7 patients with p190 bcr-abl achieved a CR, but all but one of these patients subsequently relapsed.
The observation that patients with Ph+ ALL can have evidence of p210 bcr-abl post-BMT without rapid subsequent relapse is similar to the data of CML patients post-BMT, where early PCR-positivity (<100 days post-BMT) is not predictive of subsequent relapse. Could it be that patients who express p210 were actually CML patients who were diagnosed in lymphoid blast crisis? Although this possibility is difficult to rule out completely, chart review of the patients who expressed only p210 bcr-abl did not show obvious signs of CML. Only one patient had mild splenomegaly at diagnosis, and only one (8124) had an unusual flow cytometry study which noted a small proportion of blasts that expressed myeloid markers. A leukocyte alkaline phosphatase (LAP) score was only noted in one patient and was elevated, rather than low. No BM exam at diagnosis or remission noted basophilia or a prominent left-shift of the myeloid lineage suspicious of CML.
Although there is considerable evidence demonstrating the utility of the detection of bcr-abl in CML patients post-BMT, there are few data concerning Ph+ ALL. Miyamura et al25 studied 13 patients prospectively after allogeneic (n = 8) and autologous (n = 5) transplant and found that a positive PCR assay for bcr-abl in the first 12 months after transplant was predictive of relapse. All 7 patients who were PCR-positive relapsed, whereas only 1 of 6 patients negative by PCR relapsed. Because of the small size of the study, they could not evaluate the meaning of PCR positivity in light of the possible influence of clinical variables. Moreover, because 11 of 13 patients expressed 190 bcr-abl, it was impossible to measure the influence of bcr-abl type on relapse risk.
The strength of association between a positive PCR assay and relapse found in the current study of Ph+ ALL patients is similar to that found in our previous study of ALL post-BMT using the Ig heavy chain rearrangements as the molecular fingerprint for MRD detection.37 In that study of 20 patients post-BMT, the RR of relapse associated with a positive PCR assay for MRD was 6.4.
In summary, the detection of MRD post-BMT for Ph+ ALL identifies patients at high risk for subsequent relapse. Rigorous testing for MRD post-BMT might provide a “window” in which to launch novel interventions designed to eliminate the residual leukemia before the catastrophe of rampant hematologic relapse.
Submitted August 7, 1996; accepted November 14, 1996.
Supported by National Institutes of Health Grants No. K08CA01612 and P01CA18029, and Friends of José Carreras International Leukemia Foundation.
Address reprint requests to Jerald Radich, MD, Fred Hutchinson Cancer Research Center, 1124 Columbia St, C2-023, Seattle, WA 98104.
REFERENCES
Author notes
Deceased.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal