Abstract
Both p16 and p15, encoded by the genes located on chromosome 9p21, are inhibitors of cyclin-dependent kinases (CDK4/6) and the upstream regulators of Rb function. In hematopoietic malignancies, deletion of p16/p15 locus has been shown to be highly specific to lymphoid, and more particularly from B-lineage malignancies except multiple myeloma (MM). To investigate whether these genes are inactivated by deletions, mutations, and hypermethylation of the 5′ CpG islands, we examined 12 MM patients by Southern hybridization and polymerase chain reaction–single-strand conformation polymorphism (PCR-SSCP) analysis. No deletions nor mutations of the p16 and p15 genes were found. However, hypermethylation was observed in 75% for p16 and 67% for p15 in our group of MM patients. Such high frequencies of involvement of these genes in MM make them hitherto the most common genetic abnormalities in this disease. Concomitant hypermethylation, uncommon thus far in the literature of the study of these genes, is a rather common phenomenon, occurring in 67% of our patient group. Moreover, hypermethylation of p16/p15 was associated with blastic disease and concomitant hypermethylation of both genes may be pathogenetically related to plasmacytoma development. These results indicate that these genes are important in MM pathogenesis. Here we report, for the first time in the literature, the high incidences of p16 and p15 alterations in MM, not by homozygous deletions or mutations, but solely by hypermethylation of the 5′ CpG islands, which may be a specific mechanism in this disease.
MULTIPLE MYELOMA (MM), a plasma cell tumor derived from B-lineage clonogenic cells, is a relatively rare hematologic malignancy with an elusive etiology. The age-adjusted incidence rates in the Chinese of Hong Kong per 100,000 are 1.7 for men and 1.5 for women.1 As a result of the low mitotic yield and also often complex karyotypic changes, specific cytogenetic abnormality in MM has not been found.2-5 The oncogenesis data are also rare and not systematic.6-8
Recent studies have emphasized the high incidence (50% to 70%) of hemizygous deletions of Rb using fluorescence in situ hybridization (FISH) analysis.9,10 Because Rb has a suppressive effect on the production of interleukin-6 (IL-6),11 which is a myeloma growth factor, its regulation and roles in tumor initiation and progression in MM need further evaluation.
As a negative regulator of cell cycle, Rb is subjected to the control of a broad array of cyclins, cyclin-dependent kinases, and inhibitors that play critical roles in tumorigenesis by governing balance of cell growth, division, survival, and death.12p16 and, more recently, p15, close neighbors both mapped to the 9p21 region, have been established as tumor suppressors in human neoplasias, particularly p16, which has been found homozygously deleted in multiple tumor types.13-18 Encoding, respectively, 16-kD and 15-kD proteins, they play a role in the upstream regulation of Rb function by modulating the interaction of the D1 cyclins and the CDK4/6, the complexes of which can phosphorylate and hence inactivate the Rb protein (pRb).13,19,20 The unique feature of p16 is that inactivation is mainly by homozygous deletion in primary tumors and codeletion with p15 is common.14-16 Frequent deletions of p16/p15 have been observed in lymphoid malignancies, particularly in pediatric acute lymphoblastic leukemia (ALL) and B-lineage subtype.21,22 However, thus far no deletions or mutations of p16 or p15 have been found in MM despite its B-lineage origin.22-24 Nevertheless, this may be consistent with the rarely observed loss of chromosome 9 or 9p in MM.4 6
Recently, de novo 5′ CpG island hypermethylation has been found to be associated with transcriptional silencing of the p16 and p15 genes in human cancers.17,25 Such a mechanism of p16 inactivation has also been shown to be common in many tumor types.26 27
The current study aims to investigate whether the p16/p15 genes are inactivated by deletions, mutations, or hypermethylation of the 5′ CpG islands that may serve as an alternative mechanism of inactivation of these genes in MM.
MATERIALS AND METHODS
Patients.Bone marrow (BM) materials from 12 patients diagnosed with MM in Prince of Wales Hospital, The Chinese University of Hong Kong, between May 1995 and March 1996, were recruited for the study. The diagnosis and staging classification were made following the criteria by Durie and Salmon.28-30 Morphologically, the MMs were classified into three histological subtypes, namely (1) mature (>80% of myeloma cells had mature cellular morphology); (2) intermediate (mixture of mature and immature myeloma cells present, of which 50% to 80% had mature morphology); and (3) blastic (>50% of myeloma cells were blastic with nucleolations and dissociation of nuclear and cytoplasmic maturation) through the assessment on the May-Grünwald-Giemsa–stained BM aspirate smears and hematoxylin and eosin (H & E) sections of marrow clot and trephine biopsies.
Normal controls.Twelve normal peripheral blood (PB) samples were obtained from the healthy spouses of patients with thalassemia traits as part of their premarital check. A BM aspirate sample was obtained from a healthy donor for allogeneic BM transplantation.
Southern hybridization.To assess the methylation state of p16 and p15, 5 μg of genomic DNA was first restricted with 30 U of either methylation-sensitive enzymes SacII, Sma I, or Eag I overnight as specified by the manufacturers (Amersham Corp, Buckinghamshire, UK, and Pharmacia LKB, Uppsala, Sweden). The restricted DNA was ethanol precipitated, dried, and further restricted with 30 U of EcoRI or HindIII. After fractionation on a 1% agarose gel, DNA was transferred to a Hybon N+ membrane as described by the manufacturer (Amersham Corp). Membranes were hybridized with a [32P]-labeled probe. The probe for the exon 1 of p16 was generated by the primers 2F and 1108R (Table 1), while that of the exon 1 of p15 was generated by the primers p15-1F and p15-1R (Table 1). After hybridization, the membrane was washed stringently and exposed to a Kodak X-OMAT AR film (Eastman Kodak Co, Rochester, NY) for 2 to 5 days. All cases were repeated to exclude incomplete enzymatic activity.
Primer Sequences for Analysis of p16 and p15
| Primers . | Sequences . | . | References . |
|---|---|---|---|
| M1-2F | 5′-GAAGAAAGAGGAGGGGCTG-3′ | p16, exon 1 (probe, SSCP) | Kamb et al15 |
| M1-1108R | 5′-GCGCTACCTGATTCCAATTC-3′ | p16, exon 1 (probe, SSCP) | Kamb et al15 |
| M2-42F | 5′-GGAAATTGGAAACTGGAAGC-3′ | p16, exon 2 (SSCP) | Kamb et al15 |
| M2-551R | 5′-TCTGAGCTTTGGAAGCTCT-3′ | p16, exon 2 (SSCP) | Kamb et al15 |
| X3.90F | 5′-CCGGTAGGGACGGCAAGAGA-3′ | p16, exon 3 (SSCP) | Hussussian et al31 |
| 530R | 5′-CTGTAGGACCCTCGGTGACTGATA-3′ | p16, exon 3 (SSCP) | Hussussian et al31 |
| 15 Ex 1-F | 5′-GAGGATCCGGGCCGCTGCGCGTCT-3′ | p15, exon 1 (SSCP) | Sill et al32 |
| 15 Ex 1-R | 5′-TAGGATCCAGCCCCGATCCGCCGA-3′ | p15, exon 1 (SSCP) | Sill et al32 |
| p15-1F | 5′-CCAGAAGCAATCCAGGCGCG-3′ | p15, exon 1 (probe) | Jen et al16 |
| p15-1R | 5′-AATGCACACCTCGCCAACG-3′ | p15, exon 1 (probe) | Jen et al16 |
| M2-89F | 5′-TGAGTTTAACCTGAAGGTGG-3 | p15, exon 2 (SSCP) | Kamb et al15 |
| M2-50R | 5′-GGGTGGGAAATTGGGTAAG-3 | p15, exon 2 (SSCP) | Kamb et al15 |
| Primers . | Sequences . | . | References . |
|---|---|---|---|
| M1-2F | 5′-GAAGAAAGAGGAGGGGCTG-3′ | p16, exon 1 (probe, SSCP) | Kamb et al15 |
| M1-1108R | 5′-GCGCTACCTGATTCCAATTC-3′ | p16, exon 1 (probe, SSCP) | Kamb et al15 |
| M2-42F | 5′-GGAAATTGGAAACTGGAAGC-3′ | p16, exon 2 (SSCP) | Kamb et al15 |
| M2-551R | 5′-TCTGAGCTTTGGAAGCTCT-3′ | p16, exon 2 (SSCP) | Kamb et al15 |
| X3.90F | 5′-CCGGTAGGGACGGCAAGAGA-3′ | p16, exon 3 (SSCP) | Hussussian et al31 |
| 530R | 5′-CTGTAGGACCCTCGGTGACTGATA-3′ | p16, exon 3 (SSCP) | Hussussian et al31 |
| 15 Ex 1-F | 5′-GAGGATCCGGGCCGCTGCGCGTCT-3′ | p15, exon 1 (SSCP) | Sill et al32 |
| 15 Ex 1-R | 5′-TAGGATCCAGCCCCGATCCGCCGA-3′ | p15, exon 1 (SSCP) | Sill et al32 |
| p15-1F | 5′-CCAGAAGCAATCCAGGCGCG-3′ | p15, exon 1 (probe) | Jen et al16 |
| p15-1R | 5′-AATGCACACCTCGCCAACG-3′ | p15, exon 1 (probe) | Jen et al16 |
| M2-89F | 5′-TGAGTTTAACCTGAAGGTGG-3 | p15, exon 2 (SSCP) | Kamb et al15 |
| M2-50R | 5′-GGGTGGGAAATTGGGTAAG-3 | p15, exon 2 (SSCP) | Kamb et al15 |
Polymerase chain reaction–single-strand conformation polymorphism (PCR-SSCP) analysis and direct DNA sequencing.Twelve DNA samples from MM were screened for mutations in p16 and p15 by PCR-SSCP analysis. Three exons of p16 and two exons of p15 were amplified separately as described15 using primers designed from the published sequences (Table 1). The PCR products were denatured for SSCP analysis according to Orita et al33 except that the PCR products of exon 2 of p16 and p15 were digested with Sma I before being subjected to the SSCP analysis. The PCR products that showed abnormal migration patterns on SSCP gels were reamplified, purified, and then sequenced by Delta Taq Cycle Sequencing Kit (United States Biochemical, Cleveland, OH) under conditions specified by manufacturer.
RESULTS
Patient characteristics.A total of 12 patients were analyzed. Nine patients were at stage III disease and 8 were previously untreated. Male-to-female ratio (M:F ) was 2:1, with the median age of 56 years old. The isotypic profile showed that 6 patients were of IgG, 3 IgA, 1 IgD, and 2 light-chain disease. Three patients had plasmacytomas, one from the IgG isotype and two from the light-chain disease. Histologically, 5 patients were classified as mature, 2 as intermediate, and 4 as blastic (which also included 1 patient with anaplastic features). One patient showed atypical monocytoid myeloma cells. All 12 patients had immunoparesis by protein electrophoresis. Ten patients showed lytic bone lesions. Four patients had renal failures, although none showed hypercalcamia. Ten patients had low albumins. Twenty-five percent to 100% of myeloma cells were present in the BMs through morphological assessment. Paraproteins fulfilling the major criteria by Durie were present in 8 patients, 6 of whom had it detected in the serum and 2 in the urine (Table 2). The complete blood counts showed a mean hemoglobin level of 9.6 g/dL (range, 6.9 to 11.9), white blood cell (WBC) count 4.4 × 109/L (range, 2.9 to 7.1), and platelet count 161 × 109/L (range, 52 to 432). All patients except the one with IgD myeloma were alive at the time of writing.
Clinical and Laboratory Profile of the 12 Myeloma Patients
| Patient . | Age/Sex . | Dx . | Ig . | Pcytoma . | Histology . | Stage . | R (f) . | Alb . | % BMPC . | P16 . | P15 . | Tx . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 52 M | BILPs | Gλ | − | Mature | III | N | Low | 90 | M | M | MP |
| 2* | 63 M | BILps | Dλ | − | Blastic | III | I | Low | 100 | M | M | VAD → MP |
| 3 | 42 F | BILPs | Gλ | + | Intermediate | III | N | Low | 80 | M | M | CEVAD‡ |
| 4 | 48 M | BILPs | Gλ | − | Mature | III | N | Low | 80 | N | N | MP |
| 5 | 38 M | bILPu | K | + | Monocytoid | I | N | Low | 25 | M | M | RT → CEVAD |
| 6 | 47 M | BILps | Aλ | − | Blastic† | III | N | Low | 50 | M | M | CEVAD‡ |
| 7 | 59 M | BILPs | Gλ | − | Mature | III | N | Low | 40 | N | N | MP |
| 8 | 80 F | BILps | GK | − | Mature | III | I | Low | 90 | M | M | RT + D |
| 9 | 41 F | BIPs | GK | − | Intermediate | II | N | Low | 35 | N | N | CEVAD‡ |
| 10 | 68 M | BILps | Aλ | − | Blastic | III | N | N | 52 | M | N | MP |
| 11 | 61 M | BILPu | K | + | Blastic | II | I | N | 90 | M | M | MP + RT‡ |
| 12 | 66 F | BIPs | AK | − | Mature | III | I | Low | 32 | M | M | M ± P |
| Patient . | Age/Sex . | Dx . | Ig . | Pcytoma . | Histology . | Stage . | R (f) . | Alb . | % BMPC . | P16 . | P15 . | Tx . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 52 M | BILPs | Gλ | − | Mature | III | N | Low | 90 | M | M | MP |
| 2* | 63 M | BILps | Dλ | − | Blastic | III | I | Low | 100 | M | M | VAD → MP |
| 3 | 42 F | BILPs | Gλ | + | Intermediate | III | N | Low | 80 | M | M | CEVAD‡ |
| 4 | 48 M | BILPs | Gλ | − | Mature | III | N | Low | 80 | N | N | MP |
| 5 | 38 M | bILPu | K | + | Monocytoid | I | N | Low | 25 | M | M | RT → CEVAD |
| 6 | 47 M | BILps | Aλ | − | Blastic† | III | N | Low | 50 | M | M | CEVAD‡ |
| 7 | 59 M | BILPs | Gλ | − | Mature | III | N | Low | 40 | N | N | MP |
| 8 | 80 F | BILps | GK | − | Mature | III | I | Low | 90 | M | M | RT + D |
| 9 | 41 F | BIPs | GK | − | Intermediate | II | N | Low | 35 | N | N | CEVAD‡ |
| 10 | 68 M | BILps | Aλ | − | Blastic | III | N | N | 52 | M | N | MP |
| 11 | 61 M | BILPu | K | + | Blastic | II | I | N | 90 | M | M | MP + RT‡ |
| 12 | 66 F | BIPs | AK | − | Mature | III | I | Low | 32 | M | M | M ± P |
Abbreviations: Dx, diagnosis; B, myeloma cell infiltration >30% of total nucleated cells (TNC) in BM; b, myeloma cell infiltration >10%, <30% of TNC in BM; I, immunoparesis, normal IgM <50 mg %, IgA <80 mg %, IgG <600 mg %; L, osteolytic lesions; P, major paraprotein level (fulfil major criteria of Durie & Salmon); s, serum; u, urine; p, minor paraprotein level (fulfil minor criteria of Durie & Salmon); Ig, immunoglobulin isotype; Pcytoma, plasmacytoma; R (f), renal function; N, normal; I, impaired; Alb, albumin; % BMPC, % BM plasma cells; P16, P15, M, methylation; Tx, treatment; M, melphalan; P, prednisolone; V, vincristine; A, adriamycin; D, dexamethasone; C, cyclophosphamide; E, etoposide; RT, radiotherapy.
Died shortly.
Anaplastic morphology.
Posttreated.
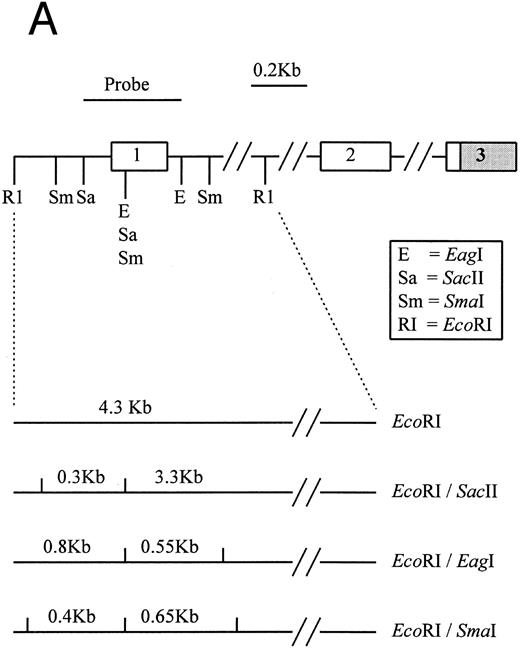
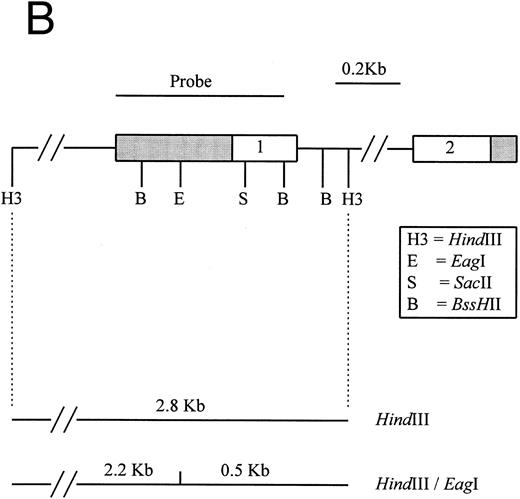
Southern blot analysis of the p16 and p15 genes with double-enzyme digestion.The presence of 5′ CpG islands located around the transcription start sites of both p16 and p15 have been described previously.17,25,26 Restriction of genomic DNA from BM aspirates of our 12 MM patients with the flanking enzyme EcoRI/HindIII, plus a methylation-sensitive enzyme (SacII, Eag I, or Sma I), on Southern hybridization with the exon 1 probes of p16 or p15 provided information on deletion and the methylation status of the 5′ CpG islands of these genes (Fig 1).17 25 As is typical of 5′ CpG island in normal cell, 12 normal PB samples and 1 normal BM aspirate sample tested showed no deletions or hypermethylation of the p16 (Fig 2A) and p15 genes. Consistent with findings in the literature, none of our MM patients had homozygous deletions of either p16 or p15 (Fig 2B and C).
(A) Restriction map of p16. Open boxes denote the three coding exons, shaded box stands for the 3′ untranslated region (UTR) of p16. Exon 1 resides in a 4.3-kb EcoRI restriction fragment. Methylation-sensitive, rare base-cutting resriction enzymes include 3 Sma I (Sm), 2 SacII (Sa), and 2 Eag I (E) sites. The location of PCR-generated probe used for Southern analysis is shown at the top. Below are the expected fragment sizes recognized by this probe when digested with the restriction enzymes shown in the normally unmethylated status. (Adapted and reprinted with permission.25 ) (B) Restriction map of p15. The probe used for Southern analysis is shown at the top. Exons 1 and 2 are depicted, with noncoding regions shaded. Methylation-sensitive restriction enzyme sites and HindIII sites are shown in the region including exon 1. The predicted sizes of restriction fragments used to analyze the methylation status of p15 are shown at the bottom. (Adapted and reprinted with permission.17 )
(A) Restriction map of p16. Open boxes denote the three coding exons, shaded box stands for the 3′ untranslated region (UTR) of p16. Exon 1 resides in a 4.3-kb EcoRI restriction fragment. Methylation-sensitive, rare base-cutting resriction enzymes include 3 Sma I (Sm), 2 SacII (Sa), and 2 Eag I (E) sites. The location of PCR-generated probe used for Southern analysis is shown at the top. Below are the expected fragment sizes recognized by this probe when digested with the restriction enzymes shown in the normally unmethylated status. (Adapted and reprinted with permission.25 ) (B) Restriction map of p15. The probe used for Southern analysis is shown at the top. Exons 1 and 2 are depicted, with noncoding regions shaded. Methylation-sensitive restriction enzyme sites and HindIII sites are shown in the region including exon 1. The predicted sizes of restriction fragments used to analyze the methylation status of p15 are shown at the bottom. (Adapted and reprinted with permission.17 )
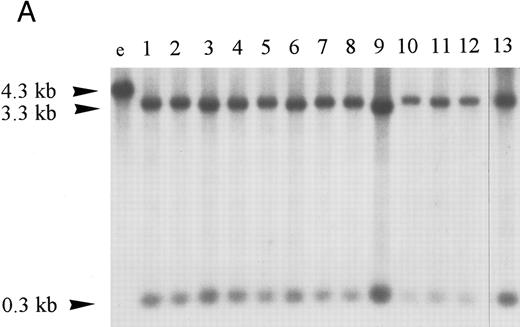
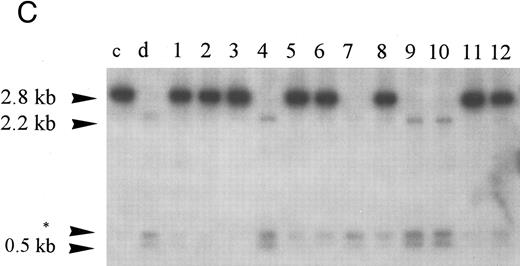
Homozygous deletion and methylation of 5′ CpG islands of p16 and p15 in MM. Lanes 1 through 12 in (B) and (C) correspond to patients no. 1 through 12 as described in the tables. (A) Southern blot analysis with the p16 exon 1 probe for normal samples (blood and BM aspirate). Lane e is normal blood DNA restricted with EcoRI alone (the 4.3-kb band simulates the fragment size seen as if the SacII sites are methylated). Lanes 1 through 12 (normal blood) and lane 13 (marrow) are DNA all restricted with EcoRI plus SacII. All lanes show no methylation band but two smaller fragments of 3.3 and 0.3 kb, which indicate absence of methylation. (B) Southern blot analysis with the p16 exon 1 probe for MM. For reference, lanes a and b are, respectively, normal blood DNA restricted with EcoRI alone (lane a, a 4.3-kb fragment simulates the fragment size seen as if the SacII sites are methylated) and EcoRI plus SacII (lane b, two smaller fragments of 3.3 and 0.3 kb, representing unmethylation status). Lanes 1 through 12 are MM DNA all restricted with EcoRI and SacII. Lanes 4, 7, and 9 show no retention of the 4.3-kb methylation bands and hence unmethylated states. All others show aberrant methylation with mixtures of 4.3-, 3.3-, and 0.3-kb fragments. (C) Southern blot analysis with the p15 exon 1 probe for MM. Similarly, lanes c and d are, respectively, normal blood DNA restricted with HindIII alone (lane c, a 2.8-kb fragment simulates the fragment size seen as if the Eag I site is methylated) and HindIII plus Eag I (lane d, two smaller fragments of 2.2 and 0.5 kb, represent unmethylation status). *An unidentified fragment recognized by the p15 exon 1 probe. Lanes 1 through 12 are MM DNA all restricted with HindIII and Eag I. Lanes 4, 7, 9, and 10 show 2.2- and 0.5-kb fragments and, hence, unmethylated states. All other lanes show the 2.8-, 2.2-, and 0.5-kb fragments and, hence, the presence of aberrant methylation.
Homozygous deletion and methylation of 5′ CpG islands of p16 and p15 in MM. Lanes 1 through 12 in (B) and (C) correspond to patients no. 1 through 12 as described in the tables. (A) Southern blot analysis with the p16 exon 1 probe for normal samples (blood and BM aspirate). Lane e is normal blood DNA restricted with EcoRI alone (the 4.3-kb band simulates the fragment size seen as if the SacII sites are methylated). Lanes 1 through 12 (normal blood) and lane 13 (marrow) are DNA all restricted with EcoRI plus SacII. All lanes show no methylation band but two smaller fragments of 3.3 and 0.3 kb, which indicate absence of methylation. (B) Southern blot analysis with the p16 exon 1 probe for MM. For reference, lanes a and b are, respectively, normal blood DNA restricted with EcoRI alone (lane a, a 4.3-kb fragment simulates the fragment size seen as if the SacII sites are methylated) and EcoRI plus SacII (lane b, two smaller fragments of 3.3 and 0.3 kb, representing unmethylation status). Lanes 1 through 12 are MM DNA all restricted with EcoRI and SacII. Lanes 4, 7, and 9 show no retention of the 4.3-kb methylation bands and hence unmethylated states. All others show aberrant methylation with mixtures of 4.3-, 3.3-, and 0.3-kb fragments. (C) Southern blot analysis with the p15 exon 1 probe for MM. Similarly, lanes c and d are, respectively, normal blood DNA restricted with HindIII alone (lane c, a 2.8-kb fragment simulates the fragment size seen as if the Eag I site is methylated) and HindIII plus Eag I (lane d, two smaller fragments of 2.2 and 0.5 kb, represent unmethylation status). *An unidentified fragment recognized by the p15 exon 1 probe. Lanes 1 through 12 are MM DNA all restricted with HindIII and Eag I. Lanes 4, 7, 9, and 10 show 2.2- and 0.5-kb fragments and, hence, unmethylated states. All other lanes show the 2.8-, 2.2-, and 0.5-kb fragments and, hence, the presence of aberrant methylation.
Hypermethylation of p16.The restriction map of p16 is as shown in Fig 1A. With 5′ CpG island hypermethylation of p16, the digestion by the methylation-sensitive enzymes would be protected and the 4.3-kb EcoRI flanking fragment would result. When the gene is unmethylated, digestion by the methylation-sensitive enzymes would yield smaller fragments. The expected sizes of these smaller fragments with different methylation-sensitive enzymes are depicted (Fig 1A). The three methylation-sensitive enzymes (SacII, Eag I, or Sma I) have different methylation-sensitive restriction sites with respect to gene sequences and, hence, locations (Fig 1A). According to Jones (1996),34 different methylation-sensitive restriction sites may be differentially methylated depending on the density and extensiveness of the methylation which varies with the developmental stage of the specific tumor. Moreover, the level of transcriptional repression is dependent on methylation density. Partial methylation of CpG islands may result in downregulation of the gene involved. With progressive methylation, the gene may be totally inactivated.34 In the 12 patients analyzed, three patients (nos. 4, 7, and 9) were unmethylated at all SacII, Eag I, and Sma I sites of p16 exon 1 (Table 3). They showed no methylation bands but two smaller fragments of 3.3 and 0.3 kb after digestion with EcoRI and SacII. Nine (75%) of our MM patients had hypermethylation at the SacII site. They all showed retention of the 4.3-kb methylation bands together with the 3.3- and 0.3-kb bands arising from the normal or unmethylated cells after digestion with EcoRI and SacII (Fig 2B). The methylation band was prominent in 5 of them (nos. 1 through 3, 11, and 12) and dominant in the other 4 (nos. 5, 6, 8, and 10) (Fig 2B). As shown from Table 3, it is noteworthy that 2 (nos. 5 and 6) of the latter samples were not methylated at the Eag I site of p16. Moreover, none of our MM samples with repeated analyses showed methylation of the Sma I site. The methylation density of the p16 gene varies among our MM patients. Taken together, these results suggest that methylation of p16 was incomplete in our MM patients.
p16 and p15 Gene Alteration in MM
| Molecular Analysis . | Methylation of p16 gene . | Methylation of p15 Gene . | Homozygous Deletion . | p16 Gene Mutation (exon 1-3) . | p15 Gene Mutation . | |||
|---|---|---|---|---|---|---|---|---|
| . | SacII . | Eag I . | Sma I . | Eag I . | p16 Gene . | p15 Gene . | . | (exon 1-2) . |
| Patient No. | ||||||||
| 1 | + | + | − | + | − | − | − | − |
| 2 | + | + | − | + | − | − | − | − |
| 3 | + | + | − | + | − | − | − | − |
| 4 | − | − | − | − | − | − | − | − |
| 5 | + | − | − | + | − | − | − | − |
| 6 | + | − | − | + | − | − | − | − |
| 7 | − | − | − | − | − | − | − | − |
| 8 | + | + | − | + | − | − | − | − |
| 9 | − | − | − | − | − | − | − | − |
| 10 | + | + | − | − | − | − | − | − |
| 11 | + | + | − | + | − | − | − | − |
| 12 | + | + | − | + | − | − | − | − |
| Molecular Analysis . | Methylation of p16 gene . | Methylation of p15 Gene . | Homozygous Deletion . | p16 Gene Mutation (exon 1-3) . | p15 Gene Mutation . | |||
|---|---|---|---|---|---|---|---|---|
| . | SacII . | Eag I . | Sma I . | Eag I . | p16 Gene . | p15 Gene . | . | (exon 1-2) . |
| Patient No. | ||||||||
| 1 | + | + | − | + | − | − | − | − |
| 2 | + | + | − | + | − | − | − | − |
| 3 | + | + | − | + | − | − | − | − |
| 4 | − | − | − | − | − | − | − | − |
| 5 | + | − | − | + | − | − | − | − |
| 6 | + | − | − | + | − | − | − | − |
| 7 | − | − | − | − | − | − | − | − |
| 8 | + | + | − | + | − | − | − | − |
| 9 | − | − | − | − | − | − | − | − |
| 10 | + | + | − | − | − | − | − | − |
| 11 | + | + | − | + | − | − | − | − |
| 12 | + | + | − | + | − | − | − | − |
Hypermethylation of p15.Similarly, the restriction map of p15 is as shown in Fig 1B. With no methylation, two smaller fragments of 2.2 and 0.5 kb would result from the digestion by HindIII and the methylation-sensitive enzyme Eag I. However, if there is 5′ CpG island methylation of p15 exon 1, a 2.8-kb restriction fragment would be produced. From the 12 patients studied, four patients (nos. 4, 7, 9, and 10) were unmethylated. All showed no methylation bands but the 2.2- and 0.5-kb fragments (Fig 2C). Eight patients (67%) had hypermethylation of p15 (Table 3). All revealed retention of the 2.8-kb methylation bands with very strong signals and the 2.2- and 0.5-kb fragments. Thus, it appears that p15 was completely methylated in all samples showing methylation.
Hypermethylation of p16/p15 and clinical pathologic correlations.We found no homozygous deletions in any of our MM patients. However, high incidences of hypermethylation were detected in p16 or p15 alone (75% and 67%, respectively) or concomitantly (67%). One patient showed only hypermethylation of p16 (no. 10) whereas 3 others were unmethylated for both genes (nos. 4, 7, and 9). Hypermethylation of p16 was found in 75% among both the pretreated (n = 8) and posttreated (n = 4) groups while that of p15 occurred in 63% among the pretreated and 75% in the posttreated groups. In the 4 patients having blastic morphology (nos. 2, 6, 10, and 11), all (100%) showed methylation of p16/p15 in contrast to the nonblastic group (n = 8) in which only 63% had hypermethylation of p16/p15. All 3 patients (100%) having plasmacytomas (nos. 3, 5, and 11) were found to have concomitant hypermethylation of both p16 and p15 genes, whereas this would only be seen in 56% of the nonplasmacytoma group (n = 9). All 3 unmethylated cases (nos. 4, 7, and 9) showed nonblastic morphology and absence of plasmacytoma (Table 2).
PCR-SSCP and DNA sequencing analysis of p16 and p15 genes.Using PCR-SSCP and DNA sequence analysis, the exons 1, 2, and 3 for p16 and exons 1 and 2 for p15 were examined. No mutations were identified in the coding sequences of these genes (Table 3).
DISCUSSION
We report for the first time in the literature that alterations of p16 and p15 occur at high incidences in MM, not by homozygous deletions or mutations, but solely by hypermethylation of 5′ CpG islands. These alterations may play an important role in the pathogenesis of MM. Other investigators have shown that hypermethylation of 5′ CpG islands of p16/p15 can lead to transcriptional silencing of the respective genes proven by the absence of transcripts.17,25,35 In vitro experiments have also shown the reversible nature of this transcriptional block by the introduction of demethylating agent, 5-aza-2′-deoxycytidine, which leads to re-expression of these genes.17,25 In our group of relatively high-stage MM patients, high frequencies of involvement of either gene alone or concomitantly were observed. However, because hypermethylation of p16/p15 was found in both low- and high-stage patients and in similar incidences in pretreated and posttreated groups, it may suggest that they are early events in MM and their role in tumor initiation can be speculated. In line with this speculation is the evidence from studies with immortal but not tumorigenic mouse cell lines, supporting the idea that methylation changes occur very early in the transformation process.34 The fact that concomitant hypermethylation of both p16 and p15 happened in all 3 cases of MM with plasmacytomas may underscore their pathogenetic significance in their development. Furthermore, the finding that all 4 MM patients with blastic morphology revealed hypermethylation of p16/p15 indicates that hypermethylation of p16/p15 may be associated with blastic disease. These are further supported by the absence of plasmacytoma and blastic morphology in all the unmethylated cases.
Both p16 and p15 are CDK4/6 inhibitors that inhibit their interaction with D1 cyclins, thus preventing the phosphorylation of the pRb and arresting the cell at G1 phase. p15 is upregulated by transforming growth factor-β (TGF-β), which plays an important role in growth suppression of hematopoietic precursor cells in the BM,36 where most of the myeloma cells would be accumulated. This growth inhibitory activity of TGF-β has been shown to be actually derived from this p15 regulation.37 In addition to p16, the inactivation of p15, which may be particularly important in BM environment where there is increased TGF-β activity,38 would thus be critical in exerting the major upstream inhibitory control of Rb function. This may also explain the need for concomitant hypermethylation of both the p16 and p15 genes in MM. The consequent loss of cell-cycle arrest and increased cellular proliferations would be further enhanced by the release of IL-6.
According to Knudson's two-hit model of tumorigenesis, both copies of tumor suppressor genes need to be inactivated.39 Although frequent deletions of Rb (50% to 70%) have been identified recently in MM, the majority are hemizygous and not associated with lack of Rb protein expression.9,10 Because biallelic loss of Rb and hence inactivation only occurs in 10% of MM,10 this may suggest that Rb is not the major target gene in this disease. Our finding of such high frequencies of hypermethylation of the p16 and p15 genes lends great support to this hypothesis. If the genes involved in the upsteam control of Rb pathway have been inactivated, further inactivation of Rb would become redundant and confer no added growth advantage for clonal selection. Examples are best seen in lung carcinomas. Although small cell lung cancers with frequent Rb inactivations rarely show p16 involvement, non–small cell lung cancers with rare Rb inactivations are more often found to have p16 inactivations.25 40 Further investigation of the Rb status in our same MM samples will be performed to study the relationship of the alterations of Rb and p16/p15.
Furthermore, if Rb is not the target gene as supported by our argument above, another possibility of frequent hemizygous Rb deletion is due to the codeletion of an unidentified tumor suppressor gene residing on the 13q14 locus, which may be another target gene for MM. Similar observations of absence of Rb inactivations have been made in some other solid tumors with frequent loss of 13q region where alternative target genes have been suggested.41 42
The problem of detection of specific gene deletions in primary tumor samples with normal cell contaminations has been addressed previously.23 25 In general, the reliability of detection decreases with increased number of normal cells. No homozygous deletions were observed in our MM patients, although it is possible that it might be an underestimation. The examination of methylation of the 5′ CpG islands of p16/p15 would be much less affected by the presence of normal cells as they would remain unmethylated and retention of a specific DNA fragment could only happen in hypermethylated genes from the tumor cells. However, the assessment of tumor infiltration on morphologic ground has potential limitation by sampling variation that may lead to a lack of correlation with the tumor contents revealed by the Southern analysis. This fact is particularly important in MM because the disease itself tends to develop focally. This sampling variation probably explains the discrepancies of tumor contents seen in some cases (eg, nos. 5 and 12). Although morphologic assessment gave both fairly low tumor percentages (25% and 32%), methylation occurred with the majority (>50%) of DNA. In these two cases, the morphologic evaluation had underestimated the tumor presence.
There are two implications for MM from our findings: (1) Because hypermethylation of p16/p15 is a common event in MM, most patients can benefit from this investigation. With PCR technique as detailed in other studies,35 43 the methylation status can serve as a tumor marker for the detection of minimal residual disease. More importantly, it may be used to check for tumor contaminations in autologous BMs or peripheral stem cell harvests in autologous BM transplantation. (2) It throws light on therapeutic innovations of MM treatment. Until now, MM was still a uniformly fatal disease with chemotherapy. Use of demethylating agents either in vitro as a pretreatment of the autologous graft before transplantation or in vivo as a therapeutic agent incorporated into drug developments or via gene therapy by targeted delivery may be speculative at this stage but effective in future.
ACKNOWLEDGMENT
The authors thank Kent Tsang for the provision of a normal BM aspirate to be included for analysis as control, and Dr Ivy Wong for review of the manuscript.
Address reprint requests to M.H.L. Ng, Associate Professor, Hematology Section, Department of Anatomical & Cellular Pathology, Prince of Wales Hospital, CUHK, Shatin, N.T., Hong Kong.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal