Abstract
We describe here the molecular basis of an isolated hereditary giant platelet disorder (GPD) which is not accompanied with thrombocytopenia or leukocyte inclusion. Platelet aggregation with ristocetin and botrocetin was almost normal in this patient. Flow cytometric analysis showed that the glycoprotein (GP) Ib/IX complex was expressed on the platelet membranes at decreased levels. The amount of platelet GPIbα and the plasma glycocalicin concentration, the water-soluble extracellular portion of GPIbα, were also decreased. The anti-GPIbα antibody coprecipitated GPIbβ and GPIX, although the ratios of these polypeptides to GPIbα was greatly decreased compared with the ratio in normal platelets. Immunoblot analysis under nonreduced conditions showed that most of the GPIbα in the patient's platelets was not disulfide linked with GPIbβ. DNA sequencing analysis showed compound heterozygosity for two independent single nucleotide substitutions: from Tyr (TAC) to Cys (TGC) at residue 88, and from Ala (GCC) to Pro (CCC) at residue 108 in her GPIbβ gene. These substitutions were not found in genomic DNA samples from 108 normal individuals. These mutations might result in decreased expression of the GPIb/IX complex and may influence the association of the complex with the membrane skeleton, consequently impairing normal platelet morphology. Furthermore, the phenotype caused by mutations in the subunits of the GPIb/IX complex could span the spectrum from a normal phenotype, to isolated GPD, to a full-blown bleeding disorder, such as Bernard-Soulier syndrome.
HEREDITARY GIANT platelet disorders (GPD) represent a rare and heterogeneous group of disorders characterized by abnormally large platelets, thrombocytopenia, and they are often associated with a tendency to bleeding.1 Well-known examples of these disorders are Bernard-Soulier syndrome (BSS)2 and the May-Hegglin anomaly.3,4 In addition, there are several forms of GPD involving nephritis and deafness.5 6 Giant platelets are also sometimes seen as an isolated finding, although the biochemical properties of giant platelets are currently unknown.
The classical diagnostic features for BSS are a prolonged bleeding time, mild to moderate thrombocytopenia, and giant platelets.2,7 Platelets from BSS patients aggregate normally to physiologic agonists such as adenosine diphosphate (ADP), epinephrine, and collagen, but fail to agglutinate with ristocetin. The defect in BSS has been identified as an absence or dysfunction of the platelet membrane glycoprotein (GP) Ib/IX complex, a platelet receptor for the von Willebrand factor (vWF ).7,8 BSS appears to be a heterogeneous syndrome not only in its clinical manifestation, but in regard to the abnormality of the GPIb/IX complex, because a residual amount of the complex and/or some of the subunits were present in some BSS patients.9-11
The GPIb/IX complex contains three polypeptide chains: the disulfide-linked α and β subunits of GPIb, and the noncovalently associated subunit, GPIX.12 The genes encoding these polypeptides have been cloned and characterized.13-15 Transfection studies showed that the complete set of three subunits are required for the efficient surface expression of a functional complex.16 These studies predicted that mutation of any one of the three subunits in vivo could cause a severe deficiency in the complex on the platelet membrane, resulting in BSS. Thus far several point mutations and a deletion mutation in the genes for GPIbα17-25 and GPIX24,26,27 have been described in BSS. Recent studies have shown that GPIbβ is the indispensable subunit linking GPIbα and GPIX.28 Incomplete complexes of GPIbβ with either GPIbα or GPIX have been demonstrated in various cell systems as well as a variant case of BSS.22,28 Recently, a small deletion in chromosome 22 that contains the GPIbβ gene was reported in a BSS patient complicated with DiGeorge syndrome.29 However, no point mutation in the GPIbβ gene has been reported. Thus, it is not known whether a mutation in GPIbβ indeed affects the synthesis and assembly of the complex in vivo.
Here we report a case with isolated GPD. The patient had no thrombocytopenia or a definite tendency to bleed. She had decreased but functional GPIb/IX complexes on her platelet membranes. The structural integrity of the complex was impaired, most likely by the disrupted disulfide linkage of GPIbα and GPIbβ. DNA sequencing analysis showed compound heterozygosity for two independent single nucleotide substitutions in the GPIbβ gene. One mutation converts Tyr (TAC) to Cys (TGC) at residue 88 and the other converts Ala (GCC) to Pro (CCC) at residue 108. These mutations appear to result in the decreased expression of the GPIb/IX complex and influence the association of the complex and the membrane skeleton, and consequently impair normal platelet morphology.
MATERIALS AND METHODS
Case history.The patient (A.K.) was a 37-year-old Japanese woman who was referred to us as probably having atypical BSS. She had giant platelets, and a normal platelet count. During childhood she had experienced frequent episodes of spontaneous epistaxis. Thereafter, her tendency to bleed settled and she has no definite spontaneous bleeding, including epistaxis and menorrhagia. Peripheral blood (PB) smears showed giant platelets with morphologically normal leukocytes. Her platelet count has been constantly normal (183 to 246 × 109/L), but the Duke's bleeding time was moderately prolonged (9.5 minutes; normal range, 2 to 5 minutes). Her platelets aggregated normally with ADP, collagen, and epinephrine, but did not with ristocetin (1.2 mg/mL). No other hematologic abnormalities were noted. Her younger sister (N.K.) was similarly affected. Her parents did not have a history of bleeding: her father had undergone total gastrectomy and her mother had given birth to four children without excessive bleeding. Her parents had no known common ancestors for at least three preceding generations.
All individuals, including the normal volunteers, gave informed consent for this study.
vWF.vWF properties were measured by vWF antigen (Ag) level with an enzyme-linked immunosorbent assay (ELISA) (Dakopatts, Copenhagen, Denmark). Ristocetin-cofactor activity (RiCof ) was determined using formalin-fixed platelets (Behringwerke, Marburg, Germany), and plasma vWF multimers were analyzed by sodium dodecyl sulfate (SDS)-agarose gel electrophoresis.30
Monoclonal and polyclonal antibodies.The anti-GPIIb/IIIa complex monoclonal antibody (MoAb) HPL1 and anti-GPIbα MoAb HPL7 have been described previously.31 The anti-GPIbβ MoAb, Gi27, was kindly provided by Dr Sentot Santoso (Justus Liebig University, Giessen, Germany). The anti-GPIX MoAb SZ1 was obtained from Immunotech SA (Marseille, France). The anti-GPV MoAb SW16 was kindly provided by Dr Piet Modderman (Netherlands Red Cross, Amsterdam, The Netherlands). The production of rabbit polyclonal antibodies directed against glycocalicin has been described previously.32 Polyclonal antiserum against GPIb/IX complex was kindly provided by Dr Makoto Handa (Keio University, Tokyo, Japan). The latter was raised in rabbits against purified GPIb/IX complex which had been purified from 5% deoxycholate-solubilized platelet membranes by a two-step procedure involving immuno-affinity chromatography with an anti-GPIbα MoAb followed by wheat-germ agglutinin-coupled agarose.
Platelet preparation.Blood samples from normal individuals, the patient, and her family members were collected into acid-citrate-dextrose (National Institutes of Health formula A). Platelet-rich plasma was prepared by centrifugation at 800g for 2 minutes. Platelets were isolated and washed three times by differential centrifugation, and resuspended in Tris-buffered saline (TBS) (10 mmol/L Tris-HCl, 0.15 mol/L NaCl, 5 mmol/L ethylenediamine tetraacetic acid, pH 7.4) containing 1 mmol/L phenylmethylsulfonyl fluoride (PMSF ) (Sigma Chemical Co, St Louis, MO) and 10 U/mL Aprotinin (Boehringer Mannheim Biochemica, Mannheim, Germany).
Ristocetin- and botrocetin-induced platelet agglutination.The blood samples were anticoagulated with 1/10 vol of 3.13% sodium citrate and platelet-rich plasma prepared by centrifugation at 800g for 2 minutes. Platelet counts were adjusted to between 200 and 250 × 109/L with autologous platelet-poor plasma. Platelet agglutination was performed in a platelet aggregometer (NBS Hema Tracer; Niko Bioscience, Tokyo, Japan). Agglutination was induced by ristocetin (Lundbeck Co, Copenhagen, Denmark) at final concentrations of 1.2, 2.0, and 2.4 mg/mL and botrocetin (a kind gift of Dr Taei Matsui, Fujita Health University, Toyoake, Japan) at concentrations of 5, 10, and 20 μg/mL.
In other experiments, platelets were washed once in TBS, resuspended in normal platelet-poor plasma, then agglutinated by adding ristocetin and botrocetin.
Flow cytometry.The washed platelet suspensions were fixed with 1% paraformaldehyde in TBS for 30 minutes at 4°C. After washing twice, the fixed platelets were incubated with 10 μg/mL of MoAbs or normal mouse IgG (Zymed Labs Inc, San Francisco, CA) for 1 hour at 4°C. The platelets were further washed and incubated with 5 μg/mL of fluorescein-conjugated F(ab)′2 fragment of goat anti-mouse IgG (Immunotech SA) for 1 hour at 4°C. Fluorescence bound to platelets was analyzed in an Epics XL flow cytometer (Coulter Electronics, Hialeah, FL).
Immunoprecipitation.Surface platelet proteins were labeled with sulfo-N-hydroxysuccinimide-biotin (NHSS-biotin; Pierce Chemical Co, Rockford, IL).33 In brief, the washed platelet suspensions were incubated with 10 mmol/L NHSS-biotin for 30 minutes at 4°C, then washed three times with Tris/glycine buffer (5 mmol/L glycine in TBS). Five micrograms of MoAb or normal mouse IgG was added to 1 mL of the labeled platelet suspension (2 × 108 platelets). After overnight incubation at 4°C, the platelets were washed twice and solubilized in lysis buffer (1% Nonidet P40 in TBS containing 1 mmol/L PMSF and 10 U/mL Aprotinin). The lysates were further incubated with 20 μL of Protein A-Sepharose CL-4B (Pharmacia Biosystems AB, Uppsala, Sweden) for 1 hour at 4°C, and the beads were sequentially washed with lysis buffer, lysis buffer containing 1 mol/L NaCl, and lysis buffer was then mixed with sample buffer containing 2% SDS and 5% 2-mercaptoethanol, and boiled for 10 minutes. The samples were separated by SDS-polyacrylamide gel electrophoresis (PAGE) on an 8% to 15% gradient acrylamide slab gel.34 The separated proteins were electroblotted onto a polyvinylidine difluoride (PVDF ) membrane (Immobilon Transfer, Millipore, Bedford, MA).35 The membrane was incubated with the avidin-horseradish peroxidase conjugate (Vector Labs Inc, Burlingame, CA), washed several times, then incubated for 1 minute with a chemiluminescent substrate (ECL Western Blotting Detection System; Amersham, Buckinghamshire, UK), then exposed to a Kodak X-Omat AR film (Eastman Kodak Co, Rochester, NY).
Immunoblot analysis.Washed platelets were solubilized in 1% Triton X-100 in TBS containing 1 mmol/L PMSF and 10 U/mL Aprotinin. They were then incubated for 30 minutes at 4°C. Solubilized platelet proteins were separated by electrophoresis on 7.5% or 12.5% SDS-PAGE slab gels. The resolved proteins were electroblotted onto PVDF membranes, incubated with the rabbit antiglycocalicin or anti-GPIb/IX complex antiserum or Gi27, washed several times, and incubated with horseradish peroxidase–conjugated goat anti-rabbit or anti-mouse antibodies (BioRad Labs, Richmond, CA). The bound antibodies were visualized using a Konica Immunostain HRP Kit (Konica, Tokyo, Japan).
Quantification of the amount of plasma glycocalicin and platelet GPIbα.The plasma glycocalicin concentration was determined by an ELISA.32 36 The total GPIbα content in Triton X-100–solubilized platelets was also measured as glycocalicin.
Polymerase chain reaction (PCR) amplification of genomic DNA.Genomic DNA was isolated from PB lymphocytes by standard procedures,37 and was applied to PCR amplification.38 Oligonucleotide primers for PCR are listed in Table 1. The target sequences were amplified with 500 ng of genomic DNA, 40 pmol of each primer, 0.2 mmol of each dNTP, and 1.25 U of cloned Pfu DNA polymerase (Stratagene, La Jolla, CA) in a total volume of 50 μL. PCR amplification proceeded in a programmable thermal cycler (model PC-700; Astec, Fukuoka, Japan) for 30 cycles of 1 minute of denaturation at 94°C, annealing for 1 minute at 60°C (for the GPIbα and vWF genes) or 61°C (for the GPIbβ and GPIX genes), and extension for 2 minutes at 74°C.
Sequences and Locations of Oligonucleotide Primers Used in the PCR Amplification
| Protein . | Primers . | Sense . | Antisense . | Location . |
|---|---|---|---|---|
| GPIbα | Ib1 | AGGTCTTTCTGCCTGCCTGT | TAGCCAGACTGAGCTTCTCC | 280-1053 |
| Ib2 | AAGGCAATGAGCTGAAGACC | CTTGTGTTGGATGCAAGGAG | 979-1575 | |
| Ib3 | TCCACTGCTTCTCTAGACAG | GGCTGATCAAGTTCAGGGAT | 1524-1960 | |
| Ib4 | CACAAGCCTGATCACTCCAA | TTCTCTCAAGGTCCCCAAAC | 1862-2496 | |
| GPIbβ | Ibb | TTACTGCGGCGCTTCCCTTG | GAGTTTGCAGGCCCGTGTTG | 641-1335 |
| Ibb3 | GCAACAACCTGACGGCGCTG | AAGGCCCAGCAGCGCAAGCT | 844-1133 | |
| GPIX | IX | TGTTCCTGCTCTGGGCCACA | TTGGTGGAGTCTGGGGACCT | 981-1560 |
| vWF | vW24 | CTGGATGTGGCGTTCGTCCT | CTCAAAGTCCTGGATGAGGATA | 4489-4965 |
| Protein . | Primers . | Sense . | Antisense . | Location . |
|---|---|---|---|---|
| GPIbα | Ib1 | AGGTCTTTCTGCCTGCCTGT | TAGCCAGACTGAGCTTCTCC | 280-1053 |
| Ib2 | AAGGCAATGAGCTGAAGACC | CTTGTGTTGGATGCAAGGAG | 979-1575 | |
| Ib3 | TCCACTGCTTCTCTAGACAG | GGCTGATCAAGTTCAGGGAT | 1524-1960 | |
| Ib4 | CACAAGCCTGATCACTCCAA | TTCTCTCAAGGTCCCCAAAC | 1862-2496 | |
| GPIbβ | Ibb | TTACTGCGGCGCTTCCCTTG | GAGTTTGCAGGCCCGTGTTG | 641-1335 |
| Ibb3 | GCAACAACCTGACGGCGCTG | AAGGCCCAGCAGCGCAAGCT | 844-1133 | |
| GPIX | IX | TGTTCCTGCTCTGGGCCACA | TTGGTGGAGTCTGGGGACCT | 981-1560 |
| vWF | vW24 | CTGGATGTGGCGTTCGTCCT | CTCAAAGTCCTGGATGAGGATA | 4489-4965 |
DNA cloning and sequencing.The DNA fragments amplified by PCR were inserted into the pCR II vector using a TA Cloning Kit (Invitrogen, San Diego, CA). The inserts were sequenced by the dideoxy nucleotide chain termination method39 using a Taq dye primer cycle sequencing kit in a model 373A DNA sequencer (Applied Biosystems, Foster City, CA).
Restriction analysis of the PCR-amplified fragments.DNA fragments were amplified using primers Ibb3, and were digested with BsoFI (New England Biolabs, Beverly, MA) or HaeIII (Boehringer Mannheim Biochemica) restriction enzymes. The digested DNA samples were electrophoresed on 15% PAGE slab gels and stained with ethidium bromide.
RESULTS
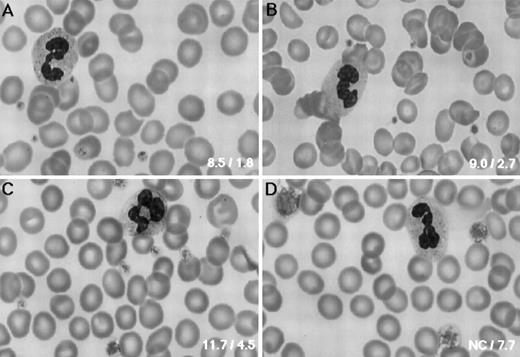
Platelet morphology.As shown in Fig 1, the PB smear of the patient revealed that she had only large platelets, which had normal morphology. Because an electronic counter (Coulter STKS; Coulter Electronics Inc) failed to determine platelet counts and the mean platelet size, these values were determined manually in a calculating chamber and on a blood smear, respectively. Her platelet counts and the mean diameter were 195 × 109/L and 7.7 μm, respectively. Her father had normal-sized platelets (2.7 μm), whereas her mother had moderately large platelets (4.5 μm) (Fig 1B and C). Her elder sister had the same size of platelets (4.5 μm) as her mother, whereas her younger sister's platelets (7.7 μm) were as large as the patient's. All individuals, including the patient, had normal platelet counts (data not shown).
Platelet morphology. PB smears were stained with May-Grünwald-Giemsa (original magnification × 800) for the normal control (A), the father (B), the mother (C), and the patient (D). The father had normal-sized platelets, whereas platelet size varied in the mother. Bottom right inset of each panel shows the platelet volume (fL) and size (μm), respectively.
Platelet morphology. PB smears were stained with May-Grünwald-Giemsa (original magnification × 800) for the normal control (A), the father (B), the mother (C), and the patient (D). The father had normal-sized platelets, whereas platelet size varied in the mother. Bottom right inset of each panel shows the platelet volume (fL) and size (μm), respectively.
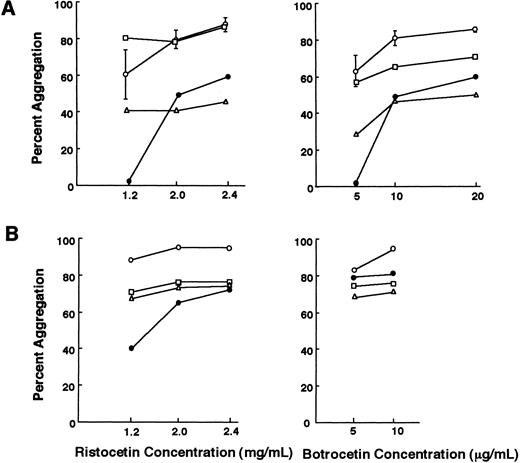
Ristocetin- and botrocetin-induced platelet agglutination.The agglutination response of platelet-rich plasma from the patient in the presence of ristocetin (1.2 mg/mL) and botrocetin (5 μg/mL) was undetectable (Fig 2A). The father had a normal platelet agglutination response, while it was slightly impaired in the mother (Fig 2A). Next we performed agglutination studies in normal platelet-poor plasma. Under this condition, the platelets from all three individuals agglutinated normally to ristocetin and botrocetin, indicating that the vWF receptor on their platelets had normal function (Fig 2B). In both experiments, the patient and her younger sister gave essentially similar results, and only the data obtained from the former are shown.
Ristocetin- and botrocetin-induced platelet agglutination. (A) PRP from the patient failed to agglutinate with ristocetin at 1.2 mg/mL and botrocetin at 5 μg/mL. The agglutination response of the mother was slightly impaired compared with that of the normal controls (n = 5). (B) Washed platelets were suspended in normal platelet-poor plasma and the platelet agglutination response was also evaluated. Platelets from the patient agglutinated to ristocetin at 1.2 mg/mL and botrocetin at 5 μg/mL. Values for the normal controls are the means ± SD. (○) Normal controls; (□) father; (▵) mother; (•) patient A.K.
Ristocetin- and botrocetin-induced platelet agglutination. (A) PRP from the patient failed to agglutinate with ristocetin at 1.2 mg/mL and botrocetin at 5 μg/mL. The agglutination response of the mother was slightly impaired compared with that of the normal controls (n = 5). (B) Washed platelets were suspended in normal platelet-poor plasma and the platelet agglutination response was also evaluated. Platelets from the patient agglutinated to ristocetin at 1.2 mg/mL and botrocetin at 5 μg/mL. Values for the normal controls are the means ± SD. (○) Normal controls; (□) father; (▵) mother; (•) patient A.K.
vWF.The patient, her younger sister, and their mother had decreased RiCof, and their RiCof/Ag ratios were also decreased. Multimeric analysis of their plasma showed an absence of the high-molecular-weight forms of vWF (Table 2). These results are compatible with patients having type 2A von Willebrand disease (vWD).
vWF Properties
| Subject . | Ag (%) . | RiCof*(%) . | Multimers . | Blood Type . |
|---|---|---|---|---|
| Father | 195 | 167 | Normal pattern | B |
| Mother | 84 | 54 | Absence of HMWM† | B |
| Patient | 86 | 50 | Absence of HMWM | B |
| Younger sister | 37 | 17 | Absence of HMWM | O |
| Subject . | Ag (%) . | RiCof*(%) . | Multimers . | Blood Type . |
|---|---|---|---|---|
| Father | 195 | 167 | Normal pattern | B |
| Mother | 84 | 54 | Absence of HMWM† | B |
| Patient | 86 | 50 | Absence of HMWM | B |
| Younger sister | 37 | 17 | Absence of HMWM | O |
Ristocetin cofactor activity.
High-molecular-weight multimers.
Because most mutations causing vWD type 2A are reported to be clustered within the A2 domain of the mature vWF subunit, between amino acid residues 742 and 875,40 we analyzed the PCR-amplified fragments encoding residues 734 to 892 in this patient. The position of primers was chosen to selectively amplify the vWF gene sequences without coamplifying the pseudogene. No mutation except some known polymorphisms was found in a total of eight independent plasmid clones.
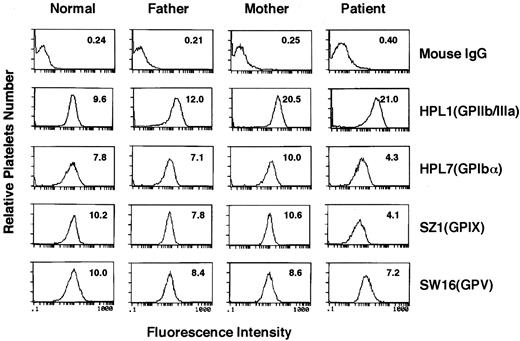
Analysis of the surface expression of GPIb/IX complex.A panel of MoAbs, directed against the GPIIb/IIIa complex (HPL1), GPIbα (HPL7), GPIX (SZ1), and GPV (SW16), was used to investigate their surface expression by flow cytometry (Fig 3). In the patient, GPIIb/IIIa complex was expressed more than in the normal control (the mean fluorescence intensity, 21.0 v 9.6), which is typical in large platelets. On the other hand, the expression of GPIbα, the GPIb/IX complex, and GPIX were remarkably lower than the normal control (4.3 v 7.8, 4.1 v 10.2, and 4.1 v 10.2, respectively). Notably, decreased but significant binding of SZ1, which recognizes GPIX combined with GPIbβ,41 indicated that GPIX formed a complex with GPIbβ on the patient's platelets. On the other hand, GPV, which is also noncovalently associated with the GPIb/IX complex, was expressed at a less decreased level than the GPIb/IX complex.
Flow cytometric analysis. Flow cytometric analysis was performed on washed, paraformaldehyde-fixed platelets from a normal individual, the patient, and her parents. The platelets were reacted with normal mouse IgG, anti-GPIIb/IIIa complex antibody (HPL1), anti-GPIbα antibody (HPL7), anti-GPIX antibody (SZ1), or anti-GPV antibody (SW16), followed by fluorescein-labeled goat anti-mouse IgG. The number in each panel indicates the mean fluorescence intensity.
Flow cytometric analysis. Flow cytometric analysis was performed on washed, paraformaldehyde-fixed platelets from a normal individual, the patient, and her parents. The platelets were reacted with normal mouse IgG, anti-GPIIb/IIIa complex antibody (HPL1), anti-GPIbα antibody (HPL7), anti-GPIX antibody (SZ1), or anti-GPV antibody (SW16), followed by fluorescein-labeled goat anti-mouse IgG. The number in each panel indicates the mean fluorescence intensity.
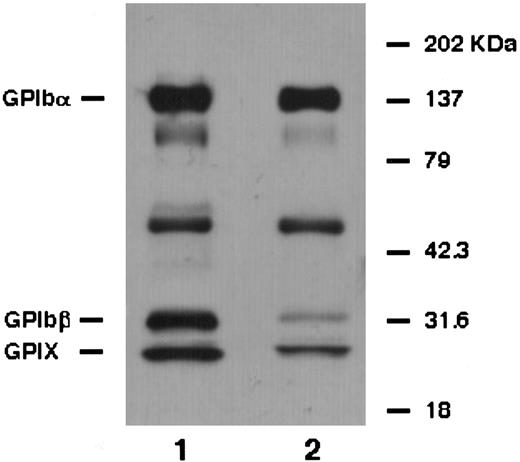
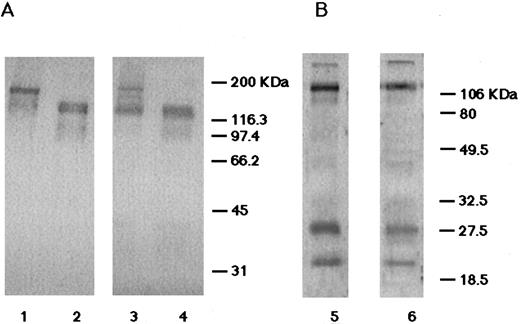
For the biochemical characterization of the GPIb/IX complex, platelets were surface-labeled and immunoprecipitated. The anti-GPIbα MoAb (HPL7) equally precipitated GPIbα, GPIbβ, and GPIX from normal platelets (Fig 4). From the patient's platelets, however, it predominantly precipitated GPIbα and a small amount of GPIbβ and GPIX (Fig 4). Moreover, the amount of GPIbβ was less than GPIX. These data suggest that the linkage between GPIbα and GPIβ was more loose than that between GPIbα and GPIX.
Immunoprecipitation analysis. Biotin-labeled platelet proteins from a normal individual (lane 1) and the patient (lane 2) were immunoprecipitated with anti-GPIbα antibody (HPL7) and analyzed under reduced conditions. From the patient's platelets, it predominantly precipitated GPIbα and a small amount of GPIbβ and GPIX. Moreover, the amount of GPIbβ was less than GPIX.
Immunoprecipitation analysis. Biotin-labeled platelet proteins from a normal individual (lane 1) and the patient (lane 2) were immunoprecipitated with anti-GPIbα antibody (HPL7) and analyzed under reduced conditions. From the patient's platelets, it predominantly precipitated GPIbα and a small amount of GPIbβ and GPIX. Moreover, the amount of GPIbβ was less than GPIX.
Detection of the GPIb/IX complex by immunoblotting.To further analyze the association of the GPIb/IX complex, solubilized whole platelet proteins were subjected to immunoblot analysis. In the normal platelets, antiglycocalicin antibodies detected 170-kD and 132-kD bands in nonreduced and reduced conditions, respectively, showing that normal GPIbα is disulfide bonded with GPIbβ. However, in the nonreduced patient's platelets, the intensity of the 170-kD band was decreased and a majority of GPIbα was detected at the molecular size of 132 kD (Fig 5A). This pattern was found even in the presence of protease inhibitors, indicating that it did not arise from the artificial degradation of GPIb. Therefore, we suspected that GPIbα and GPIbβ were not covalently linked because of the impaired disulfide bonding. Using anti-GPIb/IX complex antibodies, subnormal intensities of 27-kD (GPIbβ) and 20-kD (GPIX) bands with normal electrophoretic mobility were identified in the patient's platelets (Fig 5B).
Immunoblot analysis. (A) The immunoblots probed with the rabbit antiglycocalicin antibodies. In lanes 1 and 2, 10 μg of the solubilized normal platelets was applied. In lanes 3 and 4, 20 μg of the solubilized patient's platelets was applied. Conditions were nonreduced (lanes 1 and 2) and reduced (lanes 3 and 4). (B) The immunoblots probed with the rabbit anti-GPIb/IX complex antibodies. Lane 5, normal platelets (10 μg protein) under reduced conditions; lane 6, the patient's platelets (20 μg protein) under reduced conditions. In the patient's platelets, the amount of normal nonreduced GPIb was decreased and the majority of GPIbα was not disulfide linked with GPIbβ. GPIbβ and GPIX were identified with normal electrophoretic mobilities.
Immunoblot analysis. (A) The immunoblots probed with the rabbit antiglycocalicin antibodies. In lanes 1 and 2, 10 μg of the solubilized normal platelets was applied. In lanes 3 and 4, 20 μg of the solubilized patient's platelets was applied. Conditions were nonreduced (lanes 1 and 2) and reduced (lanes 3 and 4). (B) The immunoblots probed with the rabbit anti-GPIb/IX complex antibodies. Lane 5, normal platelets (10 μg protein) under reduced conditions; lane 6, the patient's platelets (20 μg protein) under reduced conditions. In the patient's platelets, the amount of normal nonreduced GPIb was decreased and the majority of GPIbα was not disulfide linked with GPIbβ. GPIbβ and GPIX were identified with normal electrophoretic mobilities.
GPIbβ was particularly analyzed by immunoblots probed with Gi27, anti-GPIbβ MoAb. The detected bands had the same electrophoretic mobility in all the individuals studied (data not shown), and their intensities quantified by densitometry correlated with the platelet GPIbα concentration determined by an ELISA (Table 3).
Quantification of Plasma Glycocalicin, Platelet GPIbα, and GPIbβ
| Subject . | Plasma Glycocalicin (mg/L) . | Platelet GPIbα . | Platelet GPIbβ (% normal) . |
|---|---|---|---|
| . | . | (μg/mg protein) . | . |
| Normal | 1.40 ± 0.253-150 | 9.4 ± 3.13-150 | 100 |
| Father | 1.56 (111%) | 9.5 (101%) | 86 |
| Mother | 0.84 (60%) | 5.4 (57%) | 62 |
| Patient | 0.44 (31%) | 1.9 (21%) | 28 |
| Subject . | Plasma Glycocalicin (mg/L) . | Platelet GPIbα . | Platelet GPIbβ (% normal) . |
|---|---|---|---|
| . | . | (μg/mg protein) . | . |
| Normal | 1.40 ± 0.253-150 | 9.4 ± 3.13-150 | 100 |
| Father | 1.56 (111%) | 9.5 (101%) | 86 |
| Mother | 0.84 (60%) | 5.4 (57%) | 62 |
| Patient | 0.44 (31%) | 1.9 (21%) | 28 |
The plasma glycocalicin concentration and the GPIbα concentration in platelet lysates solubilized in Triton X-100 were quantified using an ELISA. Parentheses contain the percentages of the normal values. The platelet GPIbβ content was semiquantified by densitometry of the immunoblots probed with Gi27.
The values for normal plasma glycocalicin levels and platelet GPIbα were previously reported.22
Quantification of plasma glycocalicin and platelet GPIbα.The plasma glycocalicin concentration of the patient was 0.44 mg/L (31% of normal value) and the GPIbα concentration in the platelet lysates was 1.9 μg/mg protein (21% of normal value). Her father had normal levels of plasma glycocalicin and platelet GPIbα, whereas her mother had approximately 60% of the normal values (Table 3).
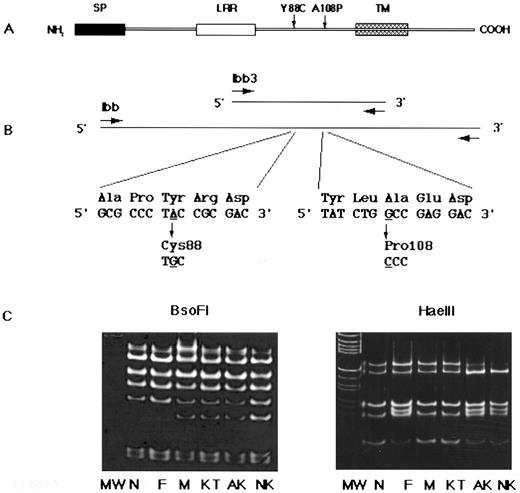
Identification of two missense mutations in the GPIbβ gene.Based on the above results, we assumed that the genetic defect(s) most probably reside on GPIbβ or in the vicinity of the GPIbβ binding region of GPIbα. Thus, we cloned and sequenced the patient's genes for the entire coding regions of GPIbβ (Fig 6A and B) and GPIbα. We found two independent single-base substitutions in her GPIbβ gene: An A to G transition at nucleotide 991, and a G to C transversion at nucleotide 1050. Of nine independent plasmid clones, six clones contained the former mutation and remaining three contained the latter. These substitutions change Tyr (TAC) to Cys (TGC) at residue 88 (Tyr88Cys), and Ala (GCC) to Pro (CCC) at residue 108 (Ala108Pro) of the mature GPIbβ protein, respectively. The Tyr88Cys mutation created an additional cleavage site for the restriction endonuclease BsoFI (GC/NGC) and that of Ala108Pro abolished a cleavage site for HaeIII (GG/CC). We took advantage of these changes and confirmed that the Tyr88Cys substitution was derived from the mother and the Ala108Pro substitution was from the father (Fig 6C). Neither substitution was found in the genomic DNA from 108 normal individuals, excluding the possibility that the mutations were polymorphisms of GPIbβ. In addition to GPIbβ, the entire coding regions of the GPIbα and GPIX genes were sequenced, but no mutations were found.
Primary structure of GPIbβ, PCR strategy, and DNA sequence analysis. (A) GPIbβ contains signal peptides (SP), a leucine-rich repeat (LRR), a transmembrane domain (TM), and a cytoplasmic tail. (B) DNA fragment amplified by the primers Ibb was cloned into the pCRII vector and sequenced. A single nucleotide substitution, an A to G transition at nucleotide 991, changed Tyr (TAC) to Cys (TGC) at residue 88 (left). A single nucleotide substitution, a G to C transversion at nucleotide 1050, changed Ala (GCC) to Pro (CCC) at residue 108 (right). Underlined nucleotides were changed. (C) Restriction analysis of the PCR-amplified GPIbβ gene. DNA fragments amplified using primers Ibb3 were digested with BsoFI or Hae III restriction enzymes, electrophoresed on 15% PAGE slab gels, and stained with ethidium bromide. The A to G transition at nucleotide 991 created a recognition site for BsoFI, generating a new 31-bp band (left). The G to C transversion at nucleotide 1050 abolished an Hae III recognition site, resulting in a new 54-bp band (right). Marker lane (M) is an Hae III digest of pBR322. MW, molecular marker; N, normal; F, father; M, mother; KT, elder sister; AK, patient; NK, younger sister.
Primary structure of GPIbβ, PCR strategy, and DNA sequence analysis. (A) GPIbβ contains signal peptides (SP), a leucine-rich repeat (LRR), a transmembrane domain (TM), and a cytoplasmic tail. (B) DNA fragment amplified by the primers Ibb was cloned into the pCRII vector and sequenced. A single nucleotide substitution, an A to G transition at nucleotide 991, changed Tyr (TAC) to Cys (TGC) at residue 88 (left). A single nucleotide substitution, a G to C transversion at nucleotide 1050, changed Ala (GCC) to Pro (CCC) at residue 108 (right). Underlined nucleotides were changed. (C) Restriction analysis of the PCR-amplified GPIbβ gene. DNA fragments amplified using primers Ibb3 were digested with BsoFI or Hae III restriction enzymes, electrophoresed on 15% PAGE slab gels, and stained with ethidium bromide. The A to G transition at nucleotide 991 created a recognition site for BsoFI, generating a new 31-bp band (left). The G to C transversion at nucleotide 1050 abolished an Hae III recognition site, resulting in a new 54-bp band (right). Marker lane (M) is an Hae III digest of pBR322. MW, molecular marker; N, normal; F, father; M, mother; KT, elder sister; AK, patient; NK, younger sister.
DISCUSSION
The aim of this study was to characterize the molecular and genetic abnormalities of an isolated case of GPD. The patient had decreased and structurally abnormal GPIb/IX complexes on her platelet membranes. The receptors retained their vWF binding properties, as demonstrated by their platelet agglutination responses to ristocetin and botrocetin. This disorder was associated with compound heterozygosity for two independent single-point mutations in the GPIbβ gene: from Tyr (TAC) to Cys (TGC) at residue 88, and from Ala (GCC) to Pro (CCC) at residue 108 in her GPIbβ gene. Several lines of evidence suggest that these two mutations are responsible for the family's platelet phenotype. First, these mutations were not polymorphisms, because they were only detected in the patient and her family members but not in 108 normal volunteers. Second, the patient and her younger sister, who both had these two mutations, and her mother and her elder sister, who both had the Tyr88Cys mutation, had identical biochemical properties of platelets. In addition, no mutations were found in the GPIbα or GPIX genes in the patient and her younger sister.
A plausible explanation for the impairment of the interchain disulfide bonds between GPIbα and GPIbβ is that the Tyr88Cys mutation in GPIbβ change the pattern of intrachain disulfide bonds without severely affecting the stability of the molecule or greatly reducing its surface expression. The three polypeptides of the complex are homologous to one another and belong to the leucine-rich glycoprotein (LRG) family.12 Although the disulfide bonding pattern for GPIbβ has not been determined, it can be drawn from homology with GPIbα and other members of the LRG family.12,42,43 Because the mutation introduces a new Cys within a probable disulfide loop, it may substitute for the more distal Cys in forming the two loops in homologous positions to those in the two polypeptides for which the disulfide pattern has been determined. This arrangement leaves a free Cys only a short distance away from the Cys postulated to bind to GPIbα. An interchain disulfide bond between these two Cys would prevent the formation of the interchain disulfide bond with GPIbα. If this hypothesis is correct, two implications about the structure of the complex can be drawn from this mutation. The first is that a residual noncovalent association may still exist between GPIbα and GPIbβ and the second is that the association between GPIbβ and GPIX is unaffected by this mutant. This second observation may also provide some insights into the associations of the polypeptides in the mature GPIb/IX complex. A recent study indicates that GPIbβ is capable of forming partial complexes with both GPIbα and GPIX.28 In that study, an association was not detected between GPIbα and GPIX when these polypeptides were expressed in various cells in the absence of GPIbβ. The GPIbβ/IX partial complex on the platelet surface was also shown in a variant case of BSS, BSS Kagoshima.22 On the other hand, another recent study suggests that in the full complex there may be a direct association between GPIbα and GPIX.44 Our data that the anti-GPIbα antibody coprecipitated GPIX to a greater degree than GPIbβ from the patient's platelets may support the presence of this association.
Another interesting feature of the platelets from individuals carrying the Tyr88Cys mutation is their large size. The abnormal size and shape of BSS platelets has been postulated to be due to the absence of a normal linkage between the platelet membrane and the cytoskeleton, a link that is mediated primarily through the GPIbα cytoplasmic domain.45 46 This explanation for the large size of BSS platelets has not been tested experimentally, but if it is correct, the Tyr88Cys mutant may indicate that the disulfide bond between GPIbα and GPIbβ is required for cytoskeletal association.
Also of interest is that despite the marked increase in platelet size, the present GPD is not associated with a decrease in platelet number, as are almost all other cases of GPD, including BSS. It is generally believed that BSS platelets are larger and more irregularly shaped because they lack the GPIb/IX complex and thus also the GPIb/IX-cytoskeleton linkage.47 Consequently, BSS platelets are considered to be much more deformable than normal platelets, and this loss of membrane stability may decrease platelet survival in the circulation, and this could account for the low platelet counts found in this disorder.48 Therefore, there is room for further investigation on the mechanisms that cause thrombocytopenia in GPD.
Where does this leave the second mutation present in these patients? Although it is difficult to make any definite conclusions about how the Ala108Pro substitution affects the structure of the polypeptide, one thing is clear: this mutation contributes to the phenotype of the two individuals who are compound heterozygotes. Two possibilities as to how it does this come to mind. First, because the Pro can change the potential of this region to form certain secondary structures, an unstable polypeptide may be formed that is degraded rapidly before being able to associate with the other two polypeptides. If the amount of GPIbβ normally synthesized is in excess to the amount of GPIbα, one might expect that the phenotype in the heterozygote would be silent because the normal allele would produce sufficient GPIbβ to complex all of the available GPIbα. This is exactly what is found in the patient's father. A second possibility is that the Ala108Pro mutant functions normally in the presence of wild-type GPIbβ (the father) but not in the presence of the Tyr88Cys mutant. This explanation requires an association on the cell surface between two GPIbβ polypeptides. This possibility is made more attractive by the recent evidence that the functional complex on the cell surface contains more than one copy of each polypeptide.49 Thus, the Ala108Pro mutant could function essentially as a dominant-negative mutant only in the presence of the other mutation. In vitro studies to express these mutants alone and in combination are in progress and should distinguish between these possibilities.
Recently, Kelly et al50 showed that a larger GPIbβ protein than in platelets is expressed in endothelial cells, and GPIbβ mRNA is expressed in the heart and brain. Although GPIbα is also present in endothelial cells, its size is identical to that observed in platelets.51 52 These findings suggest that GPIbβ may have other functional roles in other tissues. In this study, we found two missense mutations, which presumably affected platelet morphology. It is interesting whether the mutant GPIbβ protein exists in these tissues and affects their functions. However, the mutant may have no other clinical significance, because the patient is apparently healthy.
In addition to GPD, type 2A vWD was found in all the family members except the father. Although we investigated genetic basis in the A2 domain of the mature vWF subunit, where most mutations causing type 2A vWD are clustered, no mutations within this region were found. Therefore, the real cause of the decrease in the high-weight molecular forms of vWF is not known. There is a possibility, although unlikely, that the decrease in vWF in the plasma was caused by the Tyr88Cys mutation. However, if the mutant induces an abnormal increase in the binding of platelets and plasma vWF, thrombocytopenia and/or increased ristocetin- or botrocetin-induced platelet agglutination might be observed.
In summary, we have characterized the molecular lesions underlying an isolated case of GPD that is unique of a human disease caused by point mutations in the GPIbβ gene. These mutations have unexpected effects on platelet size and on the association of the GPIb/IX complex polypeptides with one another. Furthermore, our findings suggest that the phenotype caused by mutations in the subunits of the GPIb/IX complex can span the spectrum from a normal phenotype, to an isolated case of GPD, to the full-blown bleeding disorder, such as BSS. Further study of these mutations would provide insights into GPIb/IX complex biology.
Supported in part by a grant from The Naito Foundation.
Address reprint requests to Tomoki Naoe, MD, Department of Medicine, Nagoya University Branch Hospital, 1-1-20 Daiko-Minami, Higashi-ku, Nagoya 461, Japan.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal