Abstract
Several homeobox genes of the HOXA and HOXB clusters are expressed in primitive blood cells, suggesting a role for HOX genes in normal hematopoiesis. The HOXA9 gene is expressed in CD34+ marrow cells and in developing lymphocytes. We examined blood-forming organs of mice homozygous for an interrupted HOXA9 allele to determine if loss of HOX gene function is deleterious to hematopoiesis. HOXA9−/− mice have approximately 30% to 40% reductions in total leukocytes and lymphocytes (P < .001) and a blunted granulocytic response to granulocyte colony-stimulating factor (G-CSF ). Homozygous mice have significantly smaller spleens and thymuses. Myeloid/erythroid and pre-B progenitors in the marrow are significantly reduced, but no significant decreases are noted in mixed colonies, day 12 colony-forming units-spleen (CFU-S), or long-term culture–initiating cells (LTC-IC), suggesting little or no perturbation in earlier progenitors. Heterozygous animals display no hematopoietic defects. The abnormalities in leukocyte production are transplantable, indicating that the defect resides in the hematopoietic cells. These studies demonstrate a physiologic role for a HOX gene in blood cell differentiation, with the greatest apparent influence of HOXA9 at the level of the committed progenitor.
MUCH DATA HAVE emerged on the role of the HOX family of homeobox genes during embryonic development.1,2 The Hox proteins are transcription factors that have a major effect in specifying position and tissue fate in the embryo.3 Mammalian HOX genes are organized in 4 clusters (HOXA, HOXB, HOXC, and HOXD ), each containing 9 to 11 genes.4 Individual HOX genes are expressed in precise temporal and spatial patterns along various axes in the embryo,5 and targeted interruptions of HOX genes in mice often produce dramatic morphologic abnormalities of multiple tissues in specific regions of the embryo.6-8 These studies indicate a key role for HOX genes in patterning during embryogenesis.
A number of studies indicate that HOX genes are also expressed in adult tissues, including blood cells.9 Analysis of HOX gene expression in various subsets of normal marrow cells demonstrates that several members of the HOXA and HOXB loci are expressed in primitive CD34+ marrow cells, and their expression is downregulated during differentiation.10 Certain 5′ genes such as HOXA9 and HOXA10 are strongly expressed in CD34+ cells and are inactivated as cells leave the CD34+ compartment. Other more 3′ genes such as HOXB3 are downregulated within the CD34+ compartment as cells progress to the committed progenitor stage. Other studies have confirmed that HOXB3 is strongly expressed in normal marrow cells that are enriched for pluripotent progenitors, and that induction of differentiation is accompanied by an ordered 3′ to 5′ wave of HOX gene activation.11 A similar wave of HOX gene expression has been reported during T-cell activation.12 These studies suggest that sequential activation of certain HOX gene clusters may orchestrate, in part, the differentiation of various hematopoietic lineages.
A definitive physiologic role for HOX genes in blood cell development and differentiation remains to be established. Overexpression of HOXB413 or HOXB814 genes in normal murine cells results in marked expansion of primitive progenitors, suggesting that altered HOX gene expression can perturb early events in hematopoiesis. However, these observed phenotypic changes do not by themselves establish the normal function of homeobox genes in blood formation. To provide direct evidence for a role of HOX genes in the regulation of hematopoiesis, we examined the hematopoietic systems of HOXA9−/− knockout mice. These studies demonstrate that loss of function of HOXA9 results in a reduction in peripheral blood leukocytes and contractions in committed myeloid and lymphoid progenitors.
MATERIALS AND METHODS
Fluorescence-activated cell sorting, cDNA generation and amplification, and Southern blotting.Normal low-density human marrow cells were isolated, stained with directly conjugated fluorescent antibodies to CD34, CD45RA, and CD71, and then sorted on a FACStarPlus (Becton Dickinson, San Jose, CA) as described previously.10 A similar strategy was used to sort murine marrow cells and thymocytes using fluorescent antibodies to sca1, CD4, CD8, B220, CD43, IgM, and IgD. The methodology for generating and amplifying the cDNA from individual cell fractions is described in detail elsewhere.15HOXA9 expression in sorted cell populations was determined by Southern blot analysis of amplified cDNAs using 32P-labeled probes derived from the non–homeobox-containing 3′ flanking regions of the human or mouse HOXA9 gene, respectively. Total cDNA synthesis was estimated by reprobing the blots with either a human or murine β-actin probe as previously described.10
Targeting a disruption of the HOXA9 gene in murine embryonic stem cells.Mice bearing a targeted disruption of HOXA9 were furnished by Dr Cynthia Peterson and Dr Mario Capecchi. The details of engineering embryonic stem (ES) cells with a disrupted HOXA9 allele are described elsewhere (C. Peterson et al, manuscript submitted). In brief, a 1.8-kb Bg1 II/EcoRI fragment (shown in brackets in Fig 2A) of the HOXA9 gene was replaced with the MCI neo cassette. This fragment contains most of the homeobox plus 3′ noncoding sequences. The rearranged HOXA9 allele in the resulting targeted cell line 3C-3 is shown below. 3C-3 cells were injected into C57B1 blastocysts. Resultant chimeric males were crossed with C57B1/6J females to generate heterozygous offspring, which were then back-crossed to C57B1/6J mice two to four times.
HOXA9 targeting and genotypic analysis. (A) Normal HOXA9 genomic locus is shown on top and the targeted locus on the bottom. Arrowheads marked a-9#1, a-9#2, and neo denote primer sites for genotyping by PCR. (B) PCR products from tail or toe DNA, amplified using the 3 primers shown in (A). Amplification of the wild-type allele with primers a-9#1 and a-9#2 results in the lower 123-nt band, whereas amplification of the targeted allele with primers a-9#1 and neo results in the upper 200-nt band (arrows). The gel shows results from normal (lanes 1, 2, 3, and 8), HOXA9+/− (lane 4), and HOXA9−/− (lanes 5 and 6) animals. Lane 7 is a control (C) lane with no DNA.
HOXA9 targeting and genotypic analysis. (A) Normal HOXA9 genomic locus is shown on top and the targeted locus on the bottom. Arrowheads marked a-9#1, a-9#2, and neo denote primer sites for genotyping by PCR. (B) PCR products from tail or toe DNA, amplified using the 3 primers shown in (A). Amplification of the wild-type allele with primers a-9#1 and a-9#2 results in the lower 123-nt band, whereas amplification of the targeted allele with primers a-9#1 and neo results in the upper 200-nt band (arrows). The gel shows results from normal (lanes 1, 2, 3, and 8), HOXA9+/− (lane 4), and HOXA9−/− (lanes 5 and 6) animals. Lane 7 is a control (C) lane with no DNA.
Study animals and genotyping of progeny.Both HOXA9+/− and HOXA9−/− mice appear normal and are fertile. For some experiments, heterozygotes were intermated to generate homozygotes and wild-type littermates as controls. In other experiments, HOXA9−/− mice were compared with animals of the parental strains to rule out a contribution from a mixed genetic background, since the targeted 3C-3 cell line is of 129 Sv/Ev origin. In these studies, the control mice included C57B1/6J (Jackson Laboratory, Bar Harbor, ME), 129 Sv/Ev (Taconic Farms, Germantown, NY), and C57B1/129 F1 hybrid mice, which were bred and maintained at the Animal Research Facility of the Veterans Affairs Medical Center, San Francisco. HOXA9−/− animals for these experiments were generated by interbreeding homozygous mice. Mice used as marrow recipients in colony-forming units-spleen (CFU-S) assays and long-term transplantation studies were 8- to 14-week-old female C57B1/6J animals. All animals were housed in microisolator cages and provided with sterilized food and acidified water.
The progeny of various intercrosses were genotyped by polymerase chain reaction (PCR) using DNA isolated from tail or toe clips16 of weanlings and a three-primer system to distinguish wild-type from mutant HOXA9 alleles. The two HOXA9-specific primers were hoxa-9#1 (CGCTGGAACTGGAGAAGGAGTTTCTG) and hoxa-9#2 (ATCCTGCGGTTCTGGAACCAGATC); the Neo-specific primer was Neo (TCTATCGCCTTCTTGACGAGTTC) (Fig 2A). The primers hoxa-9#1 and hoxa-9#2 generate a 123-bp band representing the wild-type allele, whereas hoxa-9#1 and Neo produce a 200-bp band representing the mutant allele (Fig 2B).
Fluorescence-activated cell sorting (FACS) analysis.FACS analysis was performed using a FACScan machine (Becton Dickinson). The antibodies used included TER119 (Pharmingen, San Diego, CA) for erythroid cells; Gr-1 (Pharmingen) and Mac-1 (CalTag, South San Francisco, CA) for granulocytes and monocytes; CD3, CD4, and CD8 (CalTag) for T lymphocytes; B220 (CalTag) and CD43 (Pharmingen) for B cells; and CD34 and c-kit (Pharmingen) for primitive cells.
Assays for committed progenitors.Myeloid/erythroid progenitor assays of marrow and spleen cells from adult mice were performed in 0.8% methylcellulose in alpha medium containing 30% fetal calf serum (FCS), 3 U/mL human urinary erythropoietin, and 2% spleen cell–conditioned medium (StemCell Technologies, Vancouver, Canada) as previously described.13 To determine the distribution of progenitors, colonies from six experiments were plucked for cytospin preparations and scored for colony type. Pre-B cell colony assays of adult marrow were performed using 0.8% methylcellulose in Iscove's medium with 30% FCS, 10−4 mol/L 2-mercaptoethanol (2-ME), and 5 ng/mL interleukin-7 (Sandolfi-Winthrop, College Park, PA). Marrow cells were plated at 50,000 and 100,000 per dish, and colonies were counted at days 5 to 7. Representative colonies were plucked to confirm that the cells had lymphoid morphology.
B-lymphoid progenitor cell assay.A monolayer of the stromal cell line S1717 was established by plating 5 × 104 S17 cells on 6-cm2 tissue culture dishes in RPMI with 5% FCS and 50 mmol/L 2-ME, following previously described methods.18 When cells reached near-confluence (2 to 3 days), 1.5 × 106 unfractionated marrow cells were plated on the stromal layer. A 70% media change was performed every 3 days. After 5 weeks, nonadherent cells were washed off the dish, counted, stained with anti-B220 and anti-CD43, and evaluated by FACS analysis. B220+CD43+ cells were scored as pro-B cells, and B220+CD43− cells as pre-B cells.
Day 12 CFU-S.Marrow cells were harvested from the femora of adult HOXA9−/− mice and either wild-type littermates and C57B1 mice. Recipient C57B1 mice were lethally irradiated (800 cGy, 100 cGy/min, x-ray) and then injected with 50,000 or 100,000 unfractionated marrow cells. Untransplanted irradiated mice were tested in each experiment for endogenous CFU-S and consistently showed no spleen colonies. Twelve days after injection, animals were killed by cervical dislocation, and the number of macroscopic colonies on the spleen were evaluated after fixation in Telleyesniczky's solution.
Long-term culture–initiating cell (LTC-IC) assays.LTC-IC assays were performed as previously described.19 In brief, S17 feeder layers were established in 96-well plates, and 1 week later these were overlaid with eight different dilutions (range, .23 to 3.0 × 103 cells per well, 12 replicates of each dilution) of marrow cells harvested from adult HOXA9−/− mice and control animals. Cultures were maintained at 33°C for 4 weeks with weekly half-media exchanges. Cells were then harvested and plated in myeloid colony assays in methylcellulose, as described earlier, to determine the total clonogenic cell content of each LTC. The number of negative wells (ie, no clonogenic progenitors) was determined for each cell concentration, and the frequency of LTC-IC in the starting population was calculated using Poisson statistics and the weighted mean method.
Statistical analysis.All statistical comparisons of blood cell counts and colony numbers were performed using a two-tailed t test, using the Primer of Biostatistics Version 3.0 software for Apple Macintosh by Dr S.A. Glantz (McGraw-Hill, New York, NY). Standard errors are indicated in all tables and figures.
RESULTS
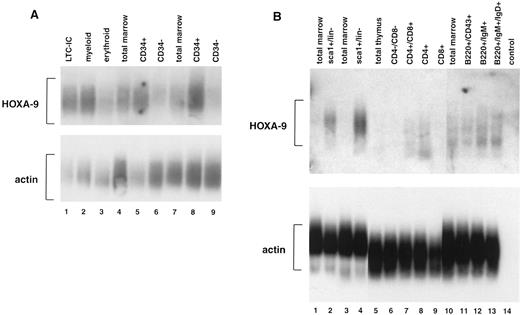
Expression of HOXA9 in normal mammalian blood cells.In previous studies using leukemic cell lines, we observed that some HOXA genes appeared to be preferentially expressed in myelomonocytic cells.20 21 To determine more accurately the expression patterns of HOXA9 in normal hematopoietic cells, we performed studies of fractionated human marrow cells using reverse transcriptase–PCR and Southern blotting, and observed that HOXA9 is expressed at high levels in CD34+ cells but only faintly if at all in CD34− cells (Fig 1A). Expression levels appear particularly high in CD34+ cells enriched for LTC-ICs (lane 1) and for myeloid progenitors (lane 2), although there is detectable message in the erythroid progenitor fraction (lane 3) as well. HOXA9 is also expressed in developing murine blood cells (Fig 1B). In sorted murine marrow, HOXA9 expression is strong in primitive sca1+/Lin− cells (lanes 2 and 4) but is not detectable in total marrow. In sorted lymphoid populations, HOXA9 message is detectable in developing lymphocytes of both T-cell (lanes 7 and 8) and B-cell (lanes 12 and 13) lineages. These studies indicate that this gene is active in normal primitive hematopoietic cells and various lymphoid populations.
Expression of HOXA9 in normal marrow cells and lymphocytes. (A) Southern blot analysis of total amplified cDNA derived from various fractions of normal human bone marrow. Lane 1, CD34+Lin− cells enriched for LTC-IC; lane 2, CD34+CD45RA+ cells enriched for myeloid progenitors; lane 3, CD34+CD71+ cells enriched for erythroid progenitors; lanes 4 and 7, total marrow cells; lanes 5 and 8, CD34+ marrow cells; lanes 6 and 9, CD34− marrow cells. HOXA9 message is clearly detectable in CD34+ cell fractions, particularly those enriched for LTC-IC and myeloid progenitors, and not in CD34− cells. (B) Southern blot analysis of total amplified cDNA from fractionated murine bone marrow, T cells, and B cells. HOXA9 is clearly detectable in primitive Sca1+/lin− marrow cells (lanes 2 and 4), but not in total marrow (lanes 1, 3, and 10). Within the T-cell fractions, there is detectable HOXA9 message in CD4+/CD8+ (lane 7) and in CD4+/CD8− (lane 8) cells. In B220+ B-cell populations, HOXA9 is expressed weakly in CD43+ cells (lane 11) and more strongly in IgM+ (lane 12) and IgM+/IgD+ (lane 13) cells.
Expression of HOXA9 in normal marrow cells and lymphocytes. (A) Southern blot analysis of total amplified cDNA derived from various fractions of normal human bone marrow. Lane 1, CD34+Lin− cells enriched for LTC-IC; lane 2, CD34+CD45RA+ cells enriched for myeloid progenitors; lane 3, CD34+CD71+ cells enriched for erythroid progenitors; lanes 4 and 7, total marrow cells; lanes 5 and 8, CD34+ marrow cells; lanes 6 and 9, CD34− marrow cells. HOXA9 message is clearly detectable in CD34+ cell fractions, particularly those enriched for LTC-IC and myeloid progenitors, and not in CD34− cells. (B) Southern blot analysis of total amplified cDNA from fractionated murine bone marrow, T cells, and B cells. HOXA9 is clearly detectable in primitive Sca1+/lin− marrow cells (lanes 2 and 4), but not in total marrow (lanes 1, 3, and 10). Within the T-cell fractions, there is detectable HOXA9 message in CD4+/CD8+ (lane 7) and in CD4+/CD8− (lane 8) cells. In B220+ B-cell populations, HOXA9 is expressed weakly in CD43+ cells (lane 11) and more strongly in IgM+ (lane 12) and IgM+/IgD+ (lane 13) cells.
Generation of HOXA9−/− mice.A targeted interruption of the HOXA9 gene was introduced into ES cells by homologous recombination as described in the methods. The resulting HOXA9+/− and HOXA9−/− mice appear entirely healthy, and weigh the same as their wild-type littermates and age-matched animals of the parental strains. Moreover, the mutant animals do not appear to be predisposed to infection or leukemia after periods of observation of more than 1 year. Figure 2B shows the genotyping of HOXA9 mutant animals using a three-primer PCR strategy. A systematic analysis of the blood and blood-forming organs of these animals was undertaken to ascertain if loss of HOXA9 function had any discernible effects on hematopoiesis.
HOXA9−/− mice have reduced numbers of peripheral blood granulocytes and lymphocytes.The white blood cell counts of homozygous animals are reduced by roughly one third compared with the counts in normal mice and heterozygous mutants, with statistically significant reductions in both lymphocytes and granulocytes (P < .001). Similar reductions in white blood cell counts are seen in HOXA9−/− mice when matched to their wild-type siblings, although the reduction in granulocytes does not achieve statistical significance (data not shown). It is notable that the HOXA9−/− animals have entirely normal red blood cell counts, hematocrits, and platelet counts (Table 1).
Peripheral Blood Cell Counts of Adult HOXA9−/−, HOXA9+/−, and Normal Mice
| Cell Type . | C57B1/6J . | HOXA9+/− . | HOXA9−/− . |
|---|---|---|---|
| . | (n = 50) . | (n = 24) . | (n = 79) . |
| Red blood cells (per μL) | 10,200,000 | 10,700,000 | 10,160,000 |
| Hematocrit (%) | 48.0 | 47.0 | 48.0 |
| Platelets (per μL) | 1,100,000 | 1,120,000 | 1,050,000 |
| White blood cells (per μL) | 9,924 ± 382 | 10,730 ± 550 | 6,850* ± 252 |
| Lymphocytes | 8,720 ± 554 | 8,988 ± 518 | 5,193* ± 405 |
| Granulocytes | 1,677 ± 124 | 1,928 ± 170 | 1,071* ± 126 |
| Cell Type . | C57B1/6J . | HOXA9+/− . | HOXA9−/− . |
|---|---|---|---|
| . | (n = 50) . | (n = 24) . | (n = 79) . |
| Red blood cells (per μL) | 10,200,000 | 10,700,000 | 10,160,000 |
| Hematocrit (%) | 48.0 | 47.0 | 48.0 |
| Platelets (per μL) | 1,100,000 | 1,120,000 | 1,050,000 |
| White blood cells (per μL) | 9,924 ± 382 | 10,730 ± 550 | 6,850* ± 252 |
| Lymphocytes | 8,720 ± 554 | 8,988 ± 518 | 5,193* ± 405 |
| Granulocytes | 1,677 ± 124 | 1,928 ± 170 | 1,071* ± 126 |
P ≤ .001 v C57B1 and HOXA9+/−.
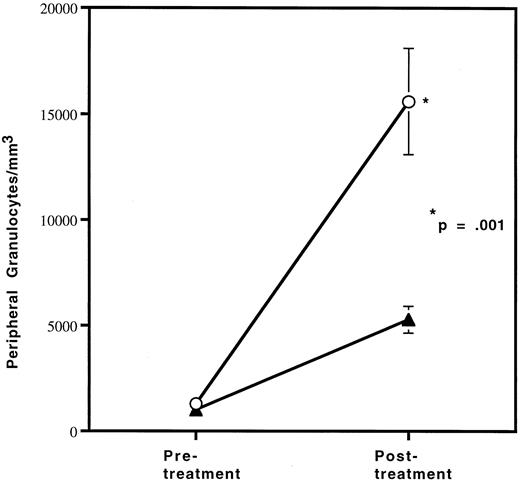
Since the reduction in granulocytes under basal conditions was small, we wished to determine if the defect in granulocyte production in HOXA9−/− mice could be exaggerated by stress. Therefore, HOXA9−/− and wild-type sibling animals were treated with granulocyte colony-stimulating factor (G-CSF ) twice daily for 96 hours and then bled for hematologic evaluation, as described previously.22 The G-CSF–induced granulocytosis in HOXA9−/− animals is only one third that seen in their wild-type siblings (P < .001) (Fig 3). Identical differences were seen in experiments comparing HOXA9−/− mice with normal C57 animals; control animals treated only with phosphate-buffered saline injections show no change in granulocyte counts (data not shown).
HOXA9−/− mice have a blunted response to G-CSF. Cohorts of (▴) 3 HOXA9−/− mice and (○) 3 HOXA9+/+ siblings were treated with twice-daily subcutaneous human G-CSF (Amgen, Thousands Oaks, CA) for 96 hours and were then bled 3 hours after the last injection. White blood cell counts were determined on a Coulter counter, and 200-cell manual differential counts of granulocytes and lymphocytes were performed on Wright-stained blood smears.
HOXA9−/− mice have a blunted response to G-CSF. Cohorts of (▴) 3 HOXA9−/− mice and (○) 3 HOXA9+/+ siblings were treated with twice-daily subcutaneous human G-CSF (Amgen, Thousands Oaks, CA) for 96 hours and were then bled 3 hours after the last injection. White blood cell counts were determined on a Coulter counter, and 200-cell manual differential counts of granulocytes and lymphocytes were performed on Wright-stained blood smears.
FACS analysis was performed to ascertain the distribution of T and B lymphocytes in HOXA9−/− mice compared with heterozygous animals and with C57B1, 129 Sv/Ev, and C57B1/129 Sv/Ev F1 hybrid animals. Figure 4 demonstrates that homozygous mutants have reductions in B220+ B cells (∼40%) compared with all the control groups (not statistically significant v 129 Sv/Ev mice) and reduction in CD3+ T cells (∼35%) compared with all the controls except C57. It should be noted that there are significant strain differences in T-/B-cell ratios between C57 and 129 Sv/Ev animals. These data demonstrate that the leukocyte abnormalities seen in the mutant animals require interruption of both alleles of HOXA9 and are not explained by differences in genetic background.
HOXA9−/− animals have reduced total white blood cell counts, granulocytes, and B and T lymphocytes. Peripheral blood cell counts were determined for (□) HOXA9 homozygotes (n = 22), () heterozygotes (n = 12), and normal parental strains including (▪) C57B1 (n = 24), (▧) 129 Sv/Ev (n = 18), and (▧) F1 hybrids of C57B1 and 129 Sv/Ev (n = 20). Animals were 9 to 12 weeks of age. WBC, total white blood cell counts; Grans, granulocytes; lymphs, lymphocytes. All counts expressed as cells/mm3 (mean ± SE). B- and T-cell counts were determined by FACS analysis using antibodies to B220 and CD3 on samples from 8 to 9 animals per group. Leukocyte counts of HOXA9−/− animals were compared with those of each group of control animals using a 2-tailed Student's t test. Control counts were significantly different from HOXA9−/− counts (P < .05), except those marked with a plus sign (+).
HOXA9−/− animals have reduced total white blood cell counts, granulocytes, and B and T lymphocytes. Peripheral blood cell counts were determined for (□) HOXA9 homozygotes (n = 22), () heterozygotes (n = 12), and normal parental strains including (▪) C57B1 (n = 24), (▧) 129 Sv/Ev (n = 18), and (▧) F1 hybrids of C57B1 and 129 Sv/Ev (n = 20). Animals were 9 to 12 weeks of age. WBC, total white blood cell counts; Grans, granulocytes; lymphs, lymphocytes. All counts expressed as cells/mm3 (mean ± SE). B- and T-cell counts were determined by FACS analysis using antibodies to B220 and CD3 on samples from 8 to 9 animals per group. Leukocyte counts of HOXA9−/− animals were compared with those of each group of control animals using a 2-tailed Student's t test. Control counts were significantly different from HOXA9−/− counts (P < .05), except those marked with a plus sign (+).
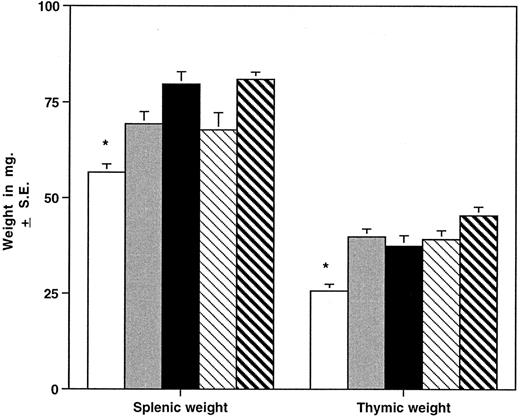
HOXA9−/− mice have smaller spleens and thymuses than normal animals.The different hematopoietic organs of the homozygous mutant mice were compared with those of heterozygous animals and normal parental strains (Fig 5). Spleens of homozygous animals are significantly smaller than those of control mice, including wild-type siblings, and grossly appear thinner and shorter in the longest diameter. The reduction in spleen size is evident in both sexes and in young (4 to 7 weeks), adult (8 to 19 weeks), and older (>20 weeks) groups and is not accounted for by a difference in the overall weight of the animals. FACS analysis of spleen cells from HOXA9−/− mice demonstrated a reduction in B cells (45% ± 7% v 61% ± 6% for C57 animals, P < .001) with a commensurate increase in the percentage of T cells (42% ± 9% v 31% ± 7% for C57, P < .01). Histologic evaluation of mutant spleens showed a reduction in white pulp and indistinct germinal centers (data not shown). These findings taken together indicate that the reduction in spleen size is largely accounted for by diminished numbers of B lymphocytes. The overall size of the thymus is also significantly reduced in HOXA9−/− mice (Fig 5); however, FACS analysis of thymic lymphocytes showed no difference in the percentages of CD4 and CD8 double- or single-positive T cells (data not shown).
HOXA9−/− mice have small spleens and thymuses. Mean ± SE splenic and thymic weights in milligrams are shown for HOXA9−/− and 4 groups of control animals. (□) A9−/− (n = 39); () A9 +/− (n = 16); (▪) C57 (n = 39); (▧) 129 (n = 26); (▧) C57/129 (n = 27). *P < .01 for A9−/− versus all other groups.
HOXA9−/− mice have small spleens and thymuses. Mean ± SE splenic and thymic weights in milligrams are shown for HOXA9−/− and 4 groups of control animals. (□) A9−/− (n = 39); () A9 +/− (n = 16); (▪) C57 (n = 39); (▧) 129 (n = 26); (▧) C57/129 (n = 27). *P < .01 for A9−/− versus all other groups.
The bone marrow of HOXA9−/−-mice displayed no evident histologic abnormalities, and the number of nucleated cells per femur is normal (mean, 17.4 ± 1.9 × 106). FACS analysis of marrow cells showed a normal 2.5:1 ratio of myeloid (Gr-1/Mac-1) to erythroid (TER119) elements and a small statistically insignificant reduction in B lymphocytes as compared with HOXA9+/+ sibling controls (21% v 27%). The percentages of CD34+ cells (7.3% v 6.4%), c-kit+ cells (7.1% v 6.8%), and double-positive cells (3.1% v 2.6%) were comparable between HOXA9−/− animals (n = 7) and their wild-type siblings (n = 5). Mesenteric lymph nodes appeared grossly normal and showed normal ratios of T and B cells (data not shown).
HOXA9−/− marrows and spleens have reduced numbers of committed myeloid and erythroid progenitors. To determine if the decrease in peripheral blood leukocytes was due to an abnormality in progenitor populations, clonogenic assays in methylcellulose were performed. A 60% reduction in total myeloid/erythroid colonies (9,600 ± 770 per/femur v 24,800 ± 3,670 per/femur, P < .001) was observed in the marrow of homozygotes (n = 8) compared with wild-type littermates (n = 6). Similar twofold reductions were seen when HOXA9−/− mice were compared with heterozygotes and all normal parental strains (data not shown).
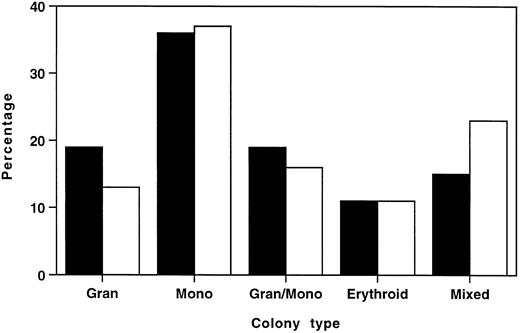
To ascertain whether this reduction in total myeloid/erythroid colonies was due to selective loss of specific colony types, individual colonies from six HOXA9−/− marrows and six normal marrows were plucked and scored by morphologic assessment of cytospin preparations. Figure 6 shows that the distribution of colony types from HOXA9−/− and normal marrow had virtually identical percentages of erythroid, monocytic, and granulocytic/monocytic colonies, and HOXA9−/− samples collectively showed a small contraction in the percentage of granulocytic colonies and an increase in the proportion of mixed erythroid/myeloid colonies. These results indicate that the decrease in total colonies was not due to selective loss of specific subsets of clonogenic progenitors, but rather to a global twofold reduction of all colony types, with the exception of mixed erythroid/myeloid colonies, which are not significantly diminished.
Distributions of myeloid/erythroid colony types in HOXA9−/− and normal marrow are similar. In 6 separate experiments, well-separated individual colonies were plucked from cultures of (□) HOXA9−/− (n = 166) and (▪) normal C57 (n = 164) marrow after 7 to 10 days of incubation. Cytospin preparations were stained and scored as 1 of 5 colony types. All colony types were represented in HOXA9−/− marrow in virtually the same percentage as in normal marrow, except for a small reduction in the percentage of pure granulocyte colonies and an increased proportion of mixed erythroid/myeloid colonies.
Distributions of myeloid/erythroid colony types in HOXA9−/− and normal marrow are similar. In 6 separate experiments, well-separated individual colonies were plucked from cultures of (□) HOXA9−/− (n = 166) and (▪) normal C57 (n = 164) marrow after 7 to 10 days of incubation. Cytospin preparations were stained and scored as 1 of 5 colony types. All colony types were represented in HOXA9−/− marrow in virtually the same percentage as in normal marrow, except for a small reduction in the percentage of pure granulocyte colonies and an increased proportion of mixed erythroid/myeloid colonies.
To identify possible defects in the proliferative potential of myeloid progenitors from HOXA9 mutant animals, whole myeloid cultures were harvested 7 to 11 days after the initial plating, and various proportions were replated for secondary colony assays. In these studies, the number of secondary colonies formed per primary colony was not reduced in HOXA9−/− animals compared with normal controls (data not shown). Finally HOXA9−/− marrow cells showed a dose-response curve to interleukin-3–induced colony formation that overlapped the curve for normal cells (data not shown). These data taken together indicate that the decrease in HOXA9−/− myeloid colony formation was not due to decreased proliferation or self-renewal or to reduced sensitivity to a hematopoietic growth factor, but rather to an absolute decrease in the number of progenitors in the marrow.
To ensure that myeloid progenitors in HOXA9−/− marrow are not simply redistributed to another hematopoietic organ, myeloid/erythroid colony assays were performed on spleen cells from HOXA9−/− and normal C57 animals. In these assays, HOXA9−/− mice showed a significant 3.5-fold reduction in total spleen-derived colonies (11.8 ± 1.6 × 103 colonies per spleen v 3.3 ± 1.2 × 103, P = .002) and a sixfold reduction in erythroid colonies (8.4 ± 1.5 × 103v 1.4 ± 0.8 × 103, P = .002), with roughly twofold reductions in granulocyte-monocyte colonies (3.1 ± 0.3 × 103v 1.7 ± 0.4 × 103, P = .02) and mixed colonies (.22 ± .11 × 103v .12 ± .04 × 103, P = .4). Similar differences were seen when mutant animals were compared with 129 Sv/Ev mice (data not shown). Thus, myeloid and erythroid progenitors were not redistributed to the spleen, and in fact, spleen-derived erythroid progenitors were markedly contracted.
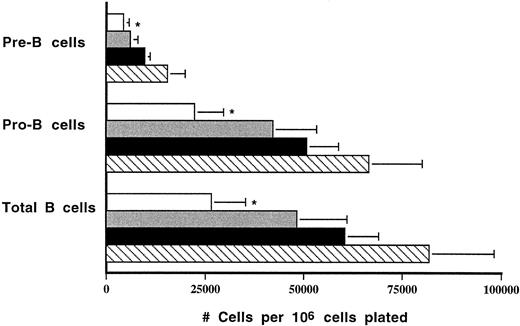
HOXA9−/− animals have defects in B-cell production.To assess B-cell progenitor frequency in the marrow, interleukin-7–responsive pre-B cell progenitors were measured, and a fourfold reduction (1,950 ± 450 calories per femur v 8,870 ± 930) was noted in HOXA9−/− marrows (n = 8) compared with wild-type sibling controls (n = 6, P < .001). Greater than twofold contractions in pre-B colonies were observed in HOXA9−/− marrows compared with marrows of heterozygotes and parental strain animals (data not shown). To measure the output of even younger B-cell precursors, bulk B-lymphoid progenitor cultures (LPC) were performed on S17 feeder layers. The output of total B cells, pro-B cells, and pre-B cells was reduced 2.5-fold to threefold in marrows of HOXA9−/− compared with both C57 and 129 Sv/Ev animals; the level of B-cell output by heterozygous mutants was intermediate between that of normal and homozygous animals (Fig 7).
Output of B cells in LPC cultures is reduced in HOXA9−/− mice. Output of B cells from 5-week LPC cultures was determined. Total B cells, B220+ cells; pro-B cells, B220+CD43+ cells; pre-B cells, B220+CD43− cells. Counts are expressed as the mean number of cells harvested at week 5 per 106 marrow cells originally plated. Marrow samples from four animals were studied in each group. (□) A9−/−; () A9+/−; (▪) C57; (▧) 129. *P < .05 for A9−/− versus C57 and 129; not significant for A9−/− versus A9+/−.
Output of B cells in LPC cultures is reduced in HOXA9−/− mice. Output of B cells from 5-week LPC cultures was determined. Total B cells, B220+ cells; pro-B cells, B220+CD43+ cells; pre-B cells, B220+CD43− cells. Counts are expressed as the mean number of cells harvested at week 5 per 106 marrow cells originally plated. Marrow samples from four animals were studied in each group. (□) A9−/−; () A9+/−; (▪) C57; (▧) 129. *P < .05 for A9−/− versus C57 and 129; not significant for A9−/− versus A9+/−.
Primitive progenitors are not significantly reduced in HOXA9−/− marrows. Having found a significant reduction in the committed progenitor population in HOXA9−/− mice, we wished to determine if there were defects earlier in the hematopoietic differentiation pathway. To measure the number of earlier multilineage precursors, day 12 CFU-S assays were performed. In two separate experiments using pooled donor marrow cells, there were small reductions in the number of CFU-S in HOXA9−/− marrows as compared with those of sibling controls or normal C57 samples (Fig 8). These differences were not significant using a two-tailed t test, although the second experiment was statistically significant using a one-tailed t test (P < .05). In one experiment injecting marrow from individual donor animals into separate cohorts of recipients, there was no difference in the number of CFU-S in HOXA9−/− and normal mice.
Day 12 CFU-S are not significantly reduced in HOXA9−/− mice. In experiments using pooled marrow samples, marrows from 2 to 3 HOXA9−/− mice were pooled and compared with pooled marrows from 2 to 3 HOXA9+/+ mice. Recipient animals (3 to 4 mice per dose of cells) were injected with either 50,000 or 100,000 pooled cells. In the experiment using individual samples, single marrow samples from 3 homozygous and 3 C57 animals were injected into cohorts of lethally irradiated C57 animals, using the same doses of cells. In 2 of 3 experiments, there was a minor and statistically insignificant reduction in the frequency of day 12 CFU-S in HOXA9−/− mice.
Day 12 CFU-S are not significantly reduced in HOXA9−/− mice. In experiments using pooled marrow samples, marrows from 2 to 3 HOXA9−/− mice were pooled and compared with pooled marrows from 2 to 3 HOXA9+/+ mice. Recipient animals (3 to 4 mice per dose of cells) were injected with either 50,000 or 100,000 pooled cells. In the experiment using individual samples, single marrow samples from 3 homozygous and 3 C57 animals were injected into cohorts of lethally irradiated C57 animals, using the same doses of cells. In 2 of 3 experiments, there was a minor and statistically insignificant reduction in the frequency of day 12 CFU-S in HOXA9−/− mice.
To assess the frequency of even earlier pluripotent progenitors, LTC-IC assays were performed. In numerous previous studies, the mean frequency of LTC-ICs in normal murine marrow was approximately one in 30,000, with 95% confidence intervals of about one in 20,000 to one in 55,000 cells. In two separate experiments, the mean LTC-IC frequency in three HOXA9−/− mice was one in 26,542, as compared with one in 36,030 for two normal mice and one in 34,051 in one heterozygote, with overlapping 95% confidence intervals for the three sets of animals. Thus, there is no evident contraction of LTC-ICs in the mutant mice.
The hematopoietic defect in HOXA9−/− mice is transplantable.The observation that HOXA9 is normally expressed in hematopoietic cells does not preclude the possibility that the hematologic defects associated with the interruption of this gene are caused at least in part by defects in the microenvironment. We determined peripheral blood cell counts of animals transplanted with either 500,000 normal C57 marrow cells or 500,000 HOXA9−/− cells. After 2 months, the hematocrits and platelet counts of the two groups were identical. However, the total white blood cell counts, granulocyte counts, and lymphocyte counts were all reduced at least twofold in animals receiving HOXA9−/− marrow, reproducing the defect seen in primary animals with somewhat greater severity. These results are consistent with a defect in leukocyte production that is intrinsic to the hematopoietic cells.
DISCUSSION
Previous studies have demonstrated that several HOX genes are expressed in early hematopoietic cells,10,11 and that overexpression of different HOX genes in normal marrow cells results in distinctive perturbations of stem cell pools and differentiation patterns.13 14 Although these studies are intriguing, they do not prove a role for HOX genes in normal hematopoiesis. We now present data indicating that the loss of function of a HOX gene normally expressed in CD34+ blood cells and in certain subsets of developing lymphocytes does disturb hematopoiesis. In the case of HOXA9, there are defects in multiple lineages, with reductions in peripheral granulocyte and lymphocyte counts and in committed myeloid, erythroid, and B-cell progenitors. The lack of anemia despite a reduction in erythroid progenitors may be due to the longer life span of mature red blood cells.
The involvement of several different lineages would suggest at first a defect in a shared progenitor. However, despite the obvious defects in committed progenitors, no major reductions were found in primitive pluripotent precursors (eg, day 12 CFU-S and LTC-IC). Although these experiments do not preclude the possibility of a mild reduction in CFU-S in HOXA9−/− marrow, they do indicate that there is not a progressively larger defect in the less mature progenitors. These findings contrast with the findings of marrow cells overexpressing the HOXB4 gene, in which case primitive progenitors are selectively and markedly expanded with little or no change in the level of committed progenitors or in the output of mature cells.13 The apparent lack of a major defect in the primitive progenitors of HOXA9−/− mice is puzzling, since HOXA9 is normally expressed in this compartment and in the committed progenitor pools (Fig 1A). It is conceivable that 3′ HOX genes such as HOXB4, which are more selectively expressed in the primitive precursors, exert a dominant effect in the earlier stages of hematopoiesis.
The contraction in progenitor pools in the HOXA9−/− mice is similar to that recently described for animals with targeted interruptions of the CD34 gene,23 although we found no change in the expression of CD34 in the marrow of HOXA9−/− mice. In both cases, it is curious that a more profound abnormality is not seen, and it is not clear if the hematopoietic defects described would confer a selection disadvantage. HOXA9−/− animals make adequate numbers of leukocytes for basal needs, but the blunted response to G-CSF suggests that these animals would be vulnerable to infectious agents in the wild.
In some other HOX gene knockout experiments, the embryologic defects have been subtle and nonlethal,24 and even examples resulting in severe abnormalities show remarkably variable penetrance even within the same animal, tempting speculation that there is redundancy in the HOX gene code.6,7 Specifically, it is conjectured that HOX gene paralogs, ie, HOX genes in the same position of different HOX clusters, such as HOXA9 and HOXB9 partially compensate for each other. In this model, one could hypothesize that the HOXB9 gene, a paralog of HOXA9 that is also expressed in blood cells,10 duplicates some of the functions of the interrupted gene. If this were the case, animals with interrupted alleles of both HOXA9 and HOXB9 may display a more severe hematopoietic defect; this is a prediction supported by the observation of combined mutations of other paralogous HOX genes, which result in much more profound skeletal defects than are seen with single mutations.25,26 There are also data to indicate that adjacent genes in the same cluster may interact to influence phenotype.27 Thus, a combined mutation of HOXA9 and its neighbor HOXA10, also strongly expressed in normal CD34+ marrow cells,10 could produce more dramatic derangements in hematopoiesis.
These observations also have relevance to recent reports of the involvement of HOXA9 in human and murine myeloid leukemias. Two groups have recently demonstrated that HOXA9 is consistently involved with the t(7; 11) translocation that occurs in a subset of human myeloid leukemias.28 29 This translocation results in fusion of the HOXA9 gene with the nucleoporin gene NUP98 and a chimeric NUP98/HOXA9 transcript, and establishes for the first time that HOX genes are involved in some spontaneous human leukemias. One group has shown that retrovirally induced myeloid leukemias in BXH-2 mice consistently involve proviral integration and activation of the murine HOXA9 gene, with resultant overexpression of HOXA9.30 In addition, we have observed that HOXA9 is consistently activated in primary samples of acute myelogenous leukemia, and not in lymphoid leukemias (H.J. Lawrence, manuscript in preparation). Taken together, these findings imply that overexpression of HOXA9 perturbs myeloid differentiation and perhaps enhances proliferation of myeloid cells, eventually leading to malignant transformation, whereas loss of HOXA9 function inhibits myeloid proliferation without markedly altering differentiation or leading to leukemia.
The nature of the hematopoietic defect in these animals remains unclear, although it is transplantable, indicating that the lesion is intrinsic to the blood cells and is not microenvironmental. The reduction in committed progenitors is apparently not due to redistribution to other hematopoietic organs or to decreased sensitivity to growth factors. A summary of the experiments performed to date examining the influence of modulating HOX gene expression on hematopoiesis would indicate that a major effect appears to be on the size of various developing blood cell pools. Alterations of pool sizes could reflect a number of aspects of cell physiology such as proliferation, survival, and apoptosis. Although much of the focus of the effects of HOX genes has been on morphogenesis, an elegant argument has been made that the role of HOX genes in body patterning is based on regulation of cell proliferation.31 An advantage of the hematopoietic system is the ease with which these cellular processes can be assayed. Thus, further studies using models of overexpression in marrow cells and analysis of the blood of knockout animals may provide insights into the general mechanisms by which HOX genes exert developmental regulatory properties.
ACKNOWLEDGMENT
We are extremely grateful to Dr Cynthia Peterson and Dr Mario Capecchi for furnishing the HOXA9−/− mice. We are also indebted to Dr Jack Levin, Georgiann Morrissey, Anne Cotleur, Dr Eva Carrier, and Erene Niemi for technical advice and assistance, and to Dr Maria Pallavicini and Dr Fern Tablin for helpful comments.
Supported by grants from the Department of Veterans Affairs (H.J.L. and C.L.), the National Cancer Institute of Canada, the Canadian Cancer Society, and the Medical Research Council (MRC) of Canada (G.S. and R.K.H.). H.J.L. is a recipient of a VA Career Development Award, C.D.H. is a recipient of a MRC postdoctoral fellowship, and G.S. is a recipient of a MRC Clinician Scientist Fellowship.
Address reprint requests to H. Jeffrey Lawrence, MD, Hematology Research (151H), Veterans Affairs Medical Center, 4150 Clement St, San Francisco, CA 94121.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal