Abstract
The D antigen is a mosaic comprising at least 30 epitopes. Partial Rh D phenotypes occur when there is absence of one or more of these epitopes, with the remainder expressed. The DVI phenotype is the most common of the partial D phenotypes, lacking most D antigen epitopes (ep D) (epD1, 2, 5-8 using the 9-epitope model or epD 1-4,7-22, 26-29 using the 30-epitope model). DVI mothers may become immunized by transfusion with D-positive blood (if typed as D-positive using polyclonal typing reagents) or by fetuses which have all of the D antigen. This situation can give rise to severe hemolytic disease of the newborn (HDN). The molecular basis of the DVI phenotype has previously been proposed to occur by two different genetic mechanisms, one (in individuals of DVICcee phenotype) where a gene conversion event generates a hybrid RHD-RHCE-RHD gene; the second (in individuals of DVIccEe phenotype) was proposed to be caused by a partial RHD gene deletion. We present evidence that in four DVICcee phenotypes studied, this phenotype is not generated by a partial RHD gene deletion, but occurs by a similar mechanism to the DVICcee phenotypes. In two individuals we have found hybrid RHD-RHCE-RHD transcripts in both DVICe and DVIcE haplotypes. These differ in that the DVICe transcripts are derived from an RHD gene where exons 4-6 have been replaced with RHCE equivalents (encoding Ala226 ); the DVIcE transcripts are derived from an RHD gene where exons 4 and 5 are replaced by RHCE equivalents (encoding Pro226 ). We provide direct evidence that Rh DVI polypeptides are expressed at the erythrocyte surface as full-length polypeptide products. We have used immunoprecipitation experiments using anti-D reactive with DVI erythrocytes followed by immunoblotting the immune complexes with rabbit sera immunoreactive to the fourth external and C-terminal domains of all Rh polypeptides. Our results illustrate that these domains are present on all Rh DVI proteins studied, and suggest that Rh DVI polypeptide species studied here exist as full-length Rh proteins.
THE HUMAN Rh D antigen is a collection of epitopes expressed by a complex composed of two different polypeptide chains, the 30-kD Rh D polypeptide and 50-kD Rh glycoprotein.1 The absence of this complex from the erythrocyte membrane, which results from a deleted2 or defective3RHD gene, causes the D-negative phenotype. Defects in the structural gene that encodes the Rh glycoprotein result in the extremely rare Rhnull phenotype, indicating that its presence during biosynthesis is critical for the assembly of all Rh proteins.4
Some D-positive individuals, initially identified by the presence of allo anti-D in their sera,5 have been classified into seven distinct categories (DI to DVII; with DI being obsolete) based on the reactivity of these antisera with red blood cells of D-variant phenotypes.6 These D-variant (or partial D) individuals lack one or more D epitopes (numbered epD1 to epD9 using the 9-epitope model7,8 or epD1 to epD30 using the 30-epitope model9 ), and thus can be immunized to them on exposure to normal D-positive erythrocytes. The DVI variant phenotype lacks most epitopes of any D-variant described (epD1, 2, 5-8 using the 9-epitope model or epD1-4, 7-22, 26-29 using the 30-epitope model), and is the most frequent of the D-variant phenotypes with gene frequencies of 0.02%, 0.02%, 0.05%, and 0.03% in English,10 United States,11 Dutch,12 and Australian13 populations, respectively. The DVI phenotype is the only known D-variant phenotype that has made a clinically significant anti-D capable of causing severe HDN and neonatal death.14 DVI phenotype erythrocytes type as D-positive when polyclonal anti-D typing reagents are used, which is problematic because DVI mothers of D-positive infants should be given postpartum anti-D.
The molecular basis of the DVI phenotype has been described previously, and is proposed to occur through two different genetic mechanisms.15 The DVICcee phenotype appears to arise through a gene conversion or double-crossing over event resulting in a hybrid RHD-RHCE-RHD gene. The hybrid DVI gene is composed of exons 1-3 of the RHD gene, 4-6 of RHCE, and 7-10 of the RHD gene. The DVIccEe phenotype is proposed to result from a partial RHD gene deletion, whereby exons 4-6 are lost. Rh D transcripts that encode a truncated 266-amino acid Rh D protein have been found, and it has been assumed that this protein, when expressed at the erythrocyte surface, is responsible for the DVIccEe phenotype.
Our findings corroborate those of Mouro et al15 concerning the DVICe phenotype. However, we present contrasting evidence concerning the DVIccEe phenotype. Our data suggest that this phenotype can occur as a result of a gene conversion event whereby exons 4 and 5 of the RHD gene are replaced by RhcE equivalents.
MATERIALS AND METHODS
Erythrocytes and antibodies.Erythrocytes of phenotype DVIccEe (donor Vi) were obtained from Dr Anna Pirkola (Finnish Red Cross Blood Transfusion Service, Helsinki, Finland). Erythrocytes from a further three individuals of DVIccEe phenotype were obtained from the Institute of Haematology, Prague; and DVICcee phenotype erythrocytes were from the Red Cross Blood Transfusion Service (Sydney, Australia) and the National Blood Service (Bristol, UK). Monoclonal antibodies (MoAbs) used were the IBGRL D typing panel (see Table 1). MoAb H41 (anti-epD3; reactive with D-positive, DVI, and DII phenotype erythrocytes) was obtained from Dr Hans Sonneborn (Biotest AG, Dreieich, Germany).
Serology of DVI Phenotype Erythrocytes
| Anti-epD . | MoAb . | Erythrocyte . | |
|---|---|---|---|
| . | . | DVICcee (8512) . | DVIccEE (Vi) . |
| epD1 | REG-A | − | − |
| epD2 | P3x249 | − | − |
| epD3 | HD7 | + | + |
| epD4 | UCHD4 | + | + |
| epD5 | HAM-A | − | − |
| epD6/7 | RUM-1 | − | − |
| epD8 | p3×21211F1 | − | − |
| epD9 | BRAD-2 | + | + |
| Anti-epD . | MoAb . | Erythrocyte . | |
|---|---|---|---|
| . | . | DVICcee (8512) . | DVIccEE (Vi) . |
| epD1 | REG-A | − | − |
| epD2 | P3x249 | − | − |
| epD3 | HD7 | + | + |
| epD4 | UCHD4 | + | + |
| epD5 | HAM-A | − | − |
| epD6/7 | RUM-1 | − | − |
| epD8 | p3×21211F1 | − | − |
| epD9 | BRAD-2 | + | + |
Positive or negative reaction profiles of two of the erythrocytes derived from DVI phenotype individuals studied (DVICe donor no. 8512; and DVIcE Vi). MoAbs used with defined specificity are shown.
Immunoblotting with rabbit antisera against Rh synthetic peptides.Rabbit sera immunoreactive with the C-terminal (anti-Ct) and fourth external domain (anti-loop 4e) of Rh polypeptides were those described previously.16,17 Immunoblotting was performed on anti-D immunoprecipitates prepared using an MoAb strongly reactive with DVI phenotype erythrocytes; H41, using the method of Moore et al.18
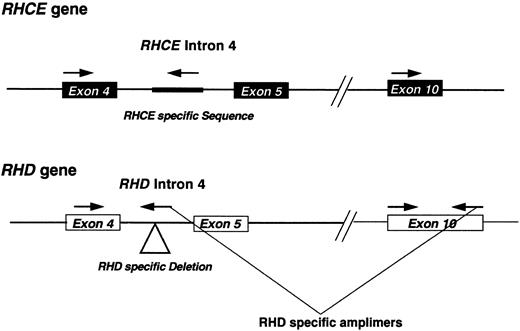
Polymerase chain reaction (PCR) Rh D typing and analysis of Rh transcripts.Genomic DNA was prepared from lymphocytes derived from DVIccEe and DVICcee phenotype individuals as described,19 and analyzed using an RHD intron 4/exon 10 multiplex assay.13,20 Briefly, the assay involves the coamplification of RHD exon 10 and intron 4, and a control RHCE intron 4 PCR product. Intron 4 RHD specificity is achieved by using a reverse PCR primer that straddles the RHD-specific deletion of this intron13,21 (Fig 1). Rh transcripts were analyzed by reverse-transcription followed by PCR (RT-PCR) using total RNA templates isolated from peripheral blood using the ammonium bicarbonate/chloride lysis method.22 PCR was performed using Bio-X-Act proof-reading DNA polymerase cocktail (Bioline, London, UK) on cDNA templates prepared using standard techniques.23 The forward primer corresponded to nucleotides (nts) −81 to −56 and had the sequence 5′-TCCCTCAAGCCCTCAAGTAG-3′. The reverse primer had the sequence 5′-GTACAAATGCAGGCAACAGTG -3′, corresponding to nts 1328-1308 of the Rh30A cDNA sequence. PCR amplification was performed using the following conditions: 94°C for 1 minute, 55°C for 1 minute, and 72°C for 3 minutes for 35 cycles in a 50-μL reaction mix composed of the following:- X1 Optiperform buffer (Bioline), 2.5 mmol/L MgCl2 1 μmol/L each primer 1.25 mmol/L each dNTP, and 2.0 U Bio-X-Act DNA polymerase cocktail. PCR products corresponding to full-length Rh transcripts were gel-purified by agarose gel electrophoresis, and cloned into pCRII vector (Invitrogen, San Diego, CA) using the manufacturer's instructions. DNA sequencing was performed on both strands of the plasmid DNA using cycle sequencing and dye-terminator chemistry on an Applied Biosystems 373A DNA sequencer (Warrington, UK).
Schematic representation of the RHD multiplex assay. A diagrammatic representation of the multiplex assay used in this study. The approximate positions of the amplimers are shown. The location of the RHD-specific deletion or RHCE-specific insertion located within intron 4 of these respective genes are highlighted on the figure.
Schematic representation of the RHD multiplex assay. A diagrammatic representation of the multiplex assay used in this study. The approximate positions of the amplimers are shown. The location of the RHD-specific deletion or RHCE-specific insertion located within intron 4 of these respective genes are highlighted on the figure.
RESULTS
Serology.The DVI phenotype was confirmed on all erythrocytes under investigation by serological testing with our panel of monoclonal anti-D to confirm the presence or absence of epitopes characteristic of this phenotype. The results obtained for DVI phenotype erythrocytes (donors 8512 [DVICcee] and Vi [DVIccEe]) are illustrated in Table 1.
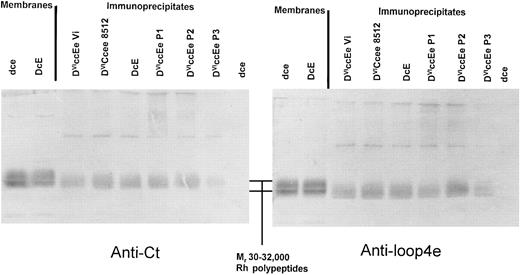
Immunoblotting of anti-D (H41; epD3) immunoprecipitates from DVICcee and DVIccEe phenotype erythrocytes. Immunoprecipitation was performed using erythrocytes obtained from four individuals expressing the DVIccEe phenotype, an individual with the DVICcee phenotype, and dce and DcE phenotypes. Immune complexes were analyzed by immunoblotting with anti-Ct and anti-loop 4e sera after separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). It was found that all immunoprecipitated Rh DVI polypeptides were of identical size (≈35 kD) and both reacted strongly with anti-Ct and anti-loop 4e (Fig 2).
Immunoprecipitation of DVICe and DVIcE proteins followed by immunoblot analysis of immune complexes with rabbit sera reactive to the fourth external and C-terminal domains of Rh polypeptides. Immunoprecipitation was performed as described in Materials and Methods, and the resultant immune complexes were separated by SDS-PAGE on 10% (wt/vol) polyacrylamide gels. Blotted membranes were then incubated with either anti-Cter or anti-loop 4e serum, and this is indicated below each panel. The position of the Rh proteins (Mr 30-32,000) is indicated in the central panel between the two blots. Control blotting samples of erythrocyte membranes are shown on the left of each immunoblot.
Immunoprecipitation of DVICe and DVIcE proteins followed by immunoblot analysis of immune complexes with rabbit sera reactive to the fourth external and C-terminal domains of Rh polypeptides. Immunoprecipitation was performed as described in Materials and Methods, and the resultant immune complexes were separated by SDS-PAGE on 10% (wt/vol) polyacrylamide gels. Blotted membranes were then incubated with either anti-Cter or anti-loop 4e serum, and this is indicated below each panel. The position of the Rh proteins (Mr 30-32,000) is indicated in the central panel between the two blots. Control blotting samples of erythrocyte membranes are shown on the left of each immunoblot.
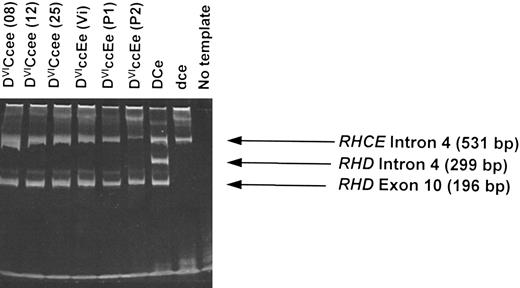
Multiplex Rh D typing of DVI genomic DNA (gDNA).Using a multiplex assay that analyzes both intron 4 and exon 10 of the RHD gene,13 we analyzed gDNA derived from 13 different individuals expressing the DVI phenotype. All samples gave a positive RHD exon 10 band but with no RHD intron 4 band while retaining the control RHCE intron 4 band (six samples illustrated in Fig 3). This is consistent with hybrid RHD-RHCE-RHD genes being present in all individuals examined. One individual (donor number P2; of phenotype DVIccEe) showed the presence of the RHD exon 10 band, but with a weak control RHCE intron 4 band (Fig 3). High-molecular-weight material in gel corresponds to gDNA template.
Multiplex RHD PCR of genomic DNA derived from DVI phenotype individuals. gDNA derived from peripheral blood lymphocytes isolated from individuals expressing DVICcee and DVIccEe phenotypes were analyzed using a RHD intron 4/exon 10 multiplex assay (see Materials and Methods). Six different individuals with the DVI phenotype are illustrated (donor identification in brackets). A D-positive (DCe), D-negative, and no template controls are also shown. PCR products were analyzed by electrophoresis on 6% polyacrylamide gels.
Multiplex RHD PCR of genomic DNA derived from DVI phenotype individuals. gDNA derived from peripheral blood lymphocytes isolated from individuals expressing DVICcee and DVIccEe phenotypes were analyzed using a RHD intron 4/exon 10 multiplex assay (see Materials and Methods). Six different individuals with the DVI phenotype are illustrated (donor identification in brackets). A D-positive (DCe), D-negative, and no template controls are also shown. PCR products were analyzed by electrophoresis on 6% polyacrylamide gels.
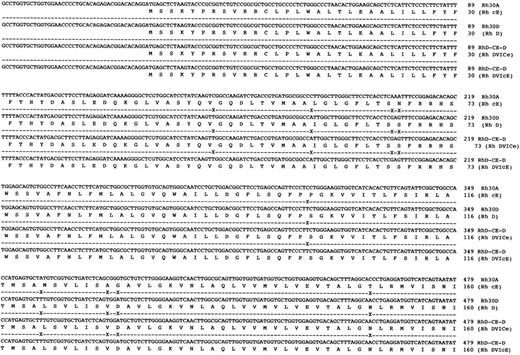
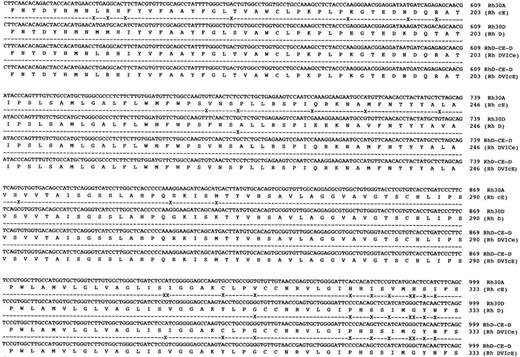
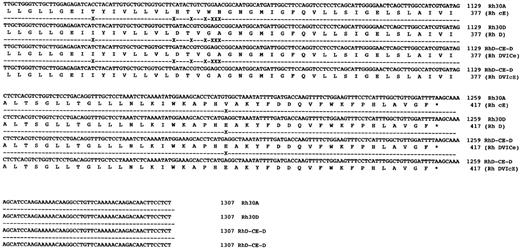
Sequence analysis of Rh transcripts derived from DVI phenotype individuals.When Rh transcripts were isolated by subcloning and sequenced (using a pool of two different gel-purified PCR reactions) from an individual expressing the DVICcee phenotype (donation no. 8512), nine clones were found to be derived from RHCE gene(s). Of these a cDNA encoding c and e antigens was fully sequenced and was found to encode an Rh ce polypeptide, and another six clones of this type were identified. Two transcripts proposed to express Rh Ce antigens were found in this individual. Another species of transcript was found, and was identical to that described by Mouro et al,15 ie, composed of RHD exons 1-3, RHCE exons 4-6, and RHD exons 7-10. Two clones of this type were sequenced fully and were found to be identical (Fig 4). Sequence analysis of full-length Rh transcripts isolated by RT-PCR from a DVIccEe phenotype individual (donor Vi) (using a pool of two different gel-purified PCR reactions) showed that Rh ce and Rh cE transcripts were present. Three Rh cE transcripts and one Rh ce transcript were sequenced fully. In addition, two identical hybrid transcripts were identified and sequenced, and were found to be composed of exons 1-3 RHD,; 4-5 RHCE, and 6-10 RHD (Fig 4). The exon 5 sequence of this hybrid transcript differed from the hybrid DVICcee transcripts inasmuch as they encoded Pro226 , the amino acid known to be critical for Rh E expression.24
DNA sequences of full-length hybrid RHD-RHCE-RHD clones isolated from DVICe and DVIcE haplotype individuals. Nucleotide sequences and inferred protein sequences are illustrated for two hybrid RHD-RHCE-RHD clones isolated by RT-PCR using total RNA derived from peripheral blood reticulocytes from two individuals expressing the DVICe and DVIcE haplotypes. Sequences are compared with RH30A and RH30D, which encode the Rh cE and D polypeptides, respectively. Differences in the Rh30D and hybrid cDNAs are denoted by (X), while sequence identity is shown by a (-).
DNA sequences of full-length hybrid RHD-RHCE-RHD clones isolated from DVICe and DVIcE haplotype individuals. Nucleotide sequences and inferred protein sequences are illustrated for two hybrid RHD-RHCE-RHD clones isolated by RT-PCR using total RNA derived from peripheral blood reticulocytes from two individuals expressing the DVICe and DVIcE haplotypes. Sequences are compared with RH30A and RH30D, which encode the Rh cE and D polypeptides, respectively. Differences in the Rh30D and hybrid cDNAs are denoted by (X), while sequence identity is shown by a (-).
DISCUSSION
This report describes the characterization of Rh transcripts and Rh DVI polypeptides in individuals expressing the Rh DVI phenotype. Although our findings confirm those of Mouro et al,15 concerning the DVICcee phenotype, we were unable to show the existence of truncated Rh D polypeptide species in individuals expressing the DVIccEe phenotype, which was proposed to occur by these workers. In recent years it has been suggested that Rh antigen expression is possible on shortened Rh polypeptide isoforms that arise by alternate splicing of Rh heterogeneous nuclear RNA (hnRNA). A model for the differential expression of Rh C or c by an alternately spliced transcript of the RHCE gene has been proposed while Rh E or e antigen expression is proposed to occur on full-length Rh polypeptides.24-26 This hypothesis is limited inasmuch as no evidence for the existence of such protein structures at the erythrocyte surface has ever been found. Two recent studies have refuted this hypothesis. Firstly, expression of Rh antigens using an ex vivo system indicates that alternate splicing is not necessary for Rh c or E expression.27 Secondly, analysis of immunopurified Rh C and c proteins indicate that they exist as full-length Rh polypeptides in the erythrocyte membrane.17 These findings indicate that Rh C or c and E or e antigen expression both occur on the same polypeptide.
It is now clear that Rh CcEe and probably DVI phenotype expression does not occur on shortened Rh isoforms. Our studies described here provide unequivocal evidence that Rh DVI proteins purified from four DVIccEe individuals studied here are not expressed on truncated Rh protein isoforms. We have shown that immunopurified Rh D proteins from all DVI phenotype erythrocytes studied express the fourth external domain of Rh proteins, which would be absent on the hypothetical shortened Rh DVIcE protein isoform (Fig 2). These proteins are of identical molecular size to other Rh polypeptides (confirming earlier findings15 ), and suggest that all Rh DVI proteins are full-length (416-amino acid) structures. We have previously shown that our SDS-PAGE analysis of Rh proteins and proteolytic fragments is sufficient to resolve amino acid differences of 50 amino acids or less,17 which refutes the hypothesis that proposed truncated Rh protein isoforms cannot be resolved by conventional SDS-PAGE. Data presented here cast some doubt as to whether the previously proposed15 truncated (266-amino acid) DVIcE protein is expressed at the erythrocyte membrane surface. The truncated protein could not be shown in any individual expressing the DVIcE haplotype in this study. The high degrees of interaction between the Rh proteins and Rh glycoprotein4 17 would indicate that disruptions to either Rh glycoprotein or Rh protein structure (in-frame stop codons, missplicing resulting in truncated polypeptide products, and/or altered reading frames) will prevent expression. It is possible that all shortened Rh transcripts described previously represent misspliced versions of Rh hnRNA primary transcripts.
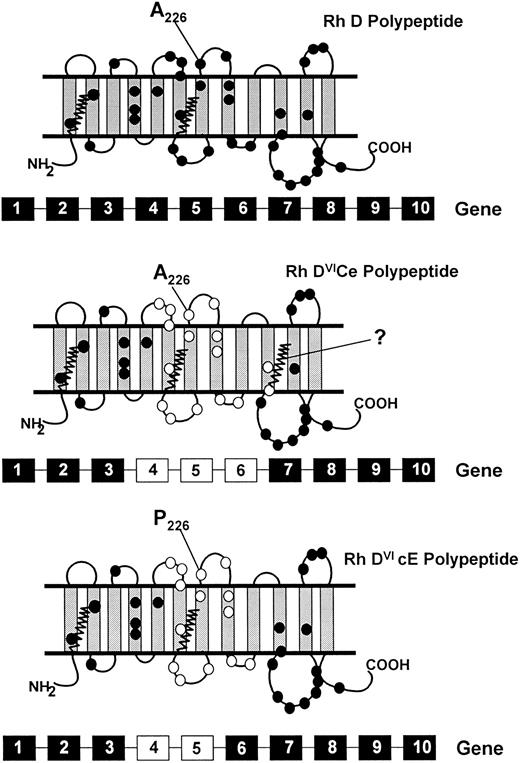
Illustration of full-length hybrid RHD-RHCE-RHD transcripts in an individual expressing the DVIccEe phenotype (donor Vi) confirms that a deleted RHD gene is not present in this individual. The Rh DVI transcripts appear to be derived from a hybrid RHD-RHCE-RHD gene comprising exons 1-3 RHD, 4-5 RHCE, and exons 6-10 RHD (Fig 5). This hybrid gene differs from that found in individuals expressing the DVICcee phenotype (exons 1-3 RHD, 4-6 RHCE, and 7-10 RHD; Fig 3). This indicates that residues encoded by RHD exons 4 and 5 are involved in epD1-2, 5-8 expression (using the 9-epitope model7,8 ) or epD1-4, 7-22, and 26-28 (using the recent 30-epitope model9 ). Exon 6 of the RHD gene encodes only two amino acid differences to the RHCE gene (Val306 and Tyr311 ), and these are likely to be located toward the internal face of the erythrocyte membrane (Fig 5). This finding indicates that RHD-specific exon 6 encoded residues are not directly involved in expression of epitopes absent from DVI phenotype erythrocytes. It is possible that other genetic mechanisms account for the DVI phenotype, but we assume that all involve rearrangement of at least exons 4 and 5 of the RHD gene, because all individuals studied were RHD intron 4-negative when typed by the multiplex assay. All DVI polypeptides are likely to be full-length protein structures similar to those depicted in Fig 5.
Schematic representation of the topologies of the Rh D, DVICe and DVIcE polypeptides. A proposed topology of the Rh D proteins is illustrated, based on hydropathy analyses31,32 and protein chemical studies.15,30 33 The Rh D protein has 35 or 36 amino acid differences to the Rh CcEe proteins, and these are highlighted with closed circles. The gene conversions that generate the DVI phenotype result in the replacement of RHD residues with RHCE counterparts. The residues involved are indicated on the DVICe and DVIcE proteins as open circles. The presence of the fourth external loop on all proteins is indicated, as well as the protein sequence difference in this domain between the DVICe and DVIcE proteins (Ala226Pro). The sites of palmitoylation (at Cys-Leu-Pro motifs) of the proteins are illustrated by zigzag lines. The genomic organization of the RHD and hybrid RHD genes are represented below each predicted topology. RHD-derived exons are represented by black boxes, RHCE-derived exons as white boxes.
Schematic representation of the topologies of the Rh D, DVICe and DVIcE polypeptides. A proposed topology of the Rh D proteins is illustrated, based on hydropathy analyses31,32 and protein chemical studies.15,30 33 The Rh D protein has 35 or 36 amino acid differences to the Rh CcEe proteins, and these are highlighted with closed circles. The gene conversions that generate the DVI phenotype result in the replacement of RHD residues with RHCE counterparts. The residues involved are indicated on the DVICe and DVIcE proteins as open circles. The presence of the fourth external loop on all proteins is indicated, as well as the protein sequence difference in this domain between the DVICe and DVIcE proteins (Ala226Pro). The sites of palmitoylation (at Cys-Leu-Pro motifs) of the proteins are illustrated by zigzag lines. The genomic organization of the RHD and hybrid RHD genes are represented below each predicted topology. RHD-derived exons are represented by black boxes, RHCE-derived exons as white boxes.
A low-frequency antigen of the Rh system, BARC (Rh52), is associated with the majority of DVICcee (a small number of DVICcee phenotype individuals are BARC-negative) but not DVIccEe phenotypes.28,29 Anti-BARC does not react with DVIccEe phenotype erythrocytes, thus sequence differences in the Rh D proteins of these two phenotypes must account for the expression of this antigen. The only exofacial difference between the DVICe and DVIcE proteins is that of residue 226, localized on the fourth external domain (Fig 5). It is possible that Ala226 (expressed by the DVICe Rh D protein, and known to be involved in Rh e antigen expression24 ), in combination with the unique external domain structure of DVI proteins, results in BARC antigen expression. Lack of BARC expression on DVIcE, Rh cE, and Rh D proteins indicate that the antigen must be discontinuous, and that the presence of Pro226 on the DVIcE protein prevents expression. Alternatively, the DVIcE Rh D protein lacks the third palmitoylation site (Cys311 -Leu-Pro) in common with normal Rh D proteins. The DVICe Rh D protein has an RHCE-specific palmitoylation site at this position (Fig 5). It is possible that conformational differences induced by differing degrees of palmitoylation results in expression of the BARC antigen.
In the United Kingdom, D-typing reagents (ie, monoclonal anti-D) are adopted to deliberately type DVI patients as D-negative. Thus, after pregnancy DVI phenotype mothers are given prophylactic anti-D. However, in instances where DVI phenotype individuals are identified as D-positive by use of inappropriate D typing reagents (eg, polyclonal anti-D), DVI mothers may become alloimmunized to their normal D-positive infants where prophylactic anti-D is not administered. In cases where alloimmunization has occurred in such cases, prenatal detection of a fetus carrying a normal RHD gene can be achieved by analysis of fetal DNA with the multiplex assay. A normal RHD intron 4–derived band would indicate that the fetus carries a paternally inherited RHD gene. The absence of the RHD intron 4–derived band and the presence of the RHD exon 10–derived band would indicate that the fetus carries only the maternally derived RhDVI gene, and would hence not be at risk of HDN. Using the multiplex assay described here, we have analyzed 357 genomic DNAs derived from D-positive, D-negative, and D-variant phenotypes.3 This study illustrates that for routine antenatal diagnosis of Rh D status, analysis of at least two regions of the RHD gene is essential: there are numerous instances where false-positive and false-negative results would be obtained when using a single RHD typing assay.
The DVIcE polypeptide does not express C antigen, but it is interesting to note that the protein is almost identical to Rh polypeptide species purported to express Rh C antigen (Fig 5; ie, Rh Ce and CE proteins). This protein encodes Ser103 , which is known to be crucial for C expression,24 but clearly not C antigen. The only exofacial differences between the DVIcE protein and Rh Ce/CE proteins are those located in the sixth external domain. It is possible that the RHCE-specific residues of this loop are essential for correct expression of C antigen. Alternatively, the DVIcE protein encodes Trp16 , whereas the Rh Ce/CE proteins all encode Cys16 , which has also been proposed to be crucial for C expression.24 The lack of Cys16 expression on the DVIcE protein may also prevent C expression. With the recent success of an in vitro system to study the expression of Rh antigens27 it is now possible to directly study the involvement of predicted critical amino acid residues in Rh antigen expression.
ACKNOWLEDGMENT
We thank Dr Geoff Daniels for useful discussion, Dr Anna Pirkola for DVIccEe phenotype erythrocytes, and Dr Hans Sonneborn for monoclonal anti-D.
Address reprint requests to Neil D. Avent, PhD, International Blood Group Reference Laboratory, Southmead Rd, Southmead, Bristol BS10 5ND, UK.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal