Abstract
Controversy exists as to whether Kaposi's sarcoma–associated herpesvirus (KSHV) is more widespread than originally reported. Recently, Monini et al reported that KSHV is ubiquitous in urogenital and prostate tissues and sperm of healthy Italian adults using nested polymerase chain reaction (PCR). We have examined for the presence of KSHV in 10 normal prostates from Italian men and 10 from men from the United States, as well as 32 prostatic, 30 vulvar, 24 ovarian, 20 cervical, and 30 testicular cancer specimens from patients from the United States. None of the patients had a history of human immunodeficiency virus infection. The samples were tested by nested PCR. The sensitivity of this assay was determined by a dilution study performed by diluting KSHV DNA from the KS-1 cells (a primary effusion lymphoma cell line which is estimated to have 16 copies of KSHV per cell) in DNA from a K562 myeloid cell line. The nested PCR that we used can detect 2.4 copies of KSHV sequences on a background of K562 DNA. All the samples were negative for KSHV sequences. Therefore, we cannot confirm the finding that KSHV sequences are ubiquitous in urogenital and prostate tissues. Furthermore, because our samples were from both the United States and Italy, the discrepancy between results is unlikely to be explained by either ethnic or environmental factors. False-positive results easily occur using nested primer PCR because of contamination. Our data argue that KSHV is not widely disseminated in urogenital tissues from nonimmunosuppressed individuals.
KAPOSI'S SARCOMA–associated herpesvirus (KSHV) or human herpesvirus 8 is a gamma-2 herpesvirus and the first member of the genus Rhadinovirus known to infect humans.1 The KSHV was identified in Kaposi's sarcoma lesions from human immunodeficiency virus–positive (HIV+) and HIV− individuals.2,3 KSHV sequences have also been detected in primary effusion lymphomas (PELs), predominantly but not exclusively in HIV+ men.4-7 Recently, we reported two cases of KSHV-associated PEL in HIV− women.8 Also, KSHV sequences have been identified in skin lesions from HIV− immunosuppressed patients,9,10 suggesting that KSHV may be more ubiquitous than originally reported. Using nested polymerase chain reaction (PCR), Monini et al11 reported that KSHV is widespread in urogenital and prostate tissues and sperm of healthy Italian adults. To elucidate the spread of KSHV infection in immunocompetent adults, we examined by nested PCR for the presence of KSHV in 10 normal prostates from Italian men and 10 from men from the United States, as well as 32 prostatic, 30 vulvar, 20 cervical, 24 ovarian, and 30 testicular cancer specimens from patients from the United States.
MATERIALS AND METHODS
Specimens were obtained from the Department of Pathology at Cedars Sinai Medical Center, Los Angeles, CA, and the Department of Medicine, Universita Cattolica, Rome, Italy. They included benign prostatic tissue from 10 men in Italy and 10 men in the United States, as well as 32 prostate carcinoma, 20 cervical carcinoma, 30 vulvar, 24 ovarian, and 30 testicular carcinoma specimens from patients in the United States.
Extraction of DNA.DNA was extracted from either frozen tissue blocks or paraffin-wax embedded tissue, as previously described.12 The quality of DNA was tested by amplifying for an endogenous gene (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]), which is present at two copies per diploid genome.13
Determination of copy numbers of KSHV in KS-1 cells.Recently, we established a cell line (KS-1) from a patient with PEL. KS-1 is infected with KSHV but not Epstein-Barr virus, cytomegalovirus, or HIV.14 KS-1 DNA was diluted in salmon sperm DNA in the ratio of 1:0, 1:1, 1:2, 1:4, 1:8, 1:16, 1:32, and 1:64, respectively. The diluted DNA were amplified with primers which amplify open reading frame (ORF ) 73 of KSHV, primer: 5′-GCAGTCTCCAGAGTCTTCTC-3′,3′ primer: 5′-CGGAGCTAAAGAGTCTGGTG-3′,15 and GAPDH, primer: 5′-CCACCATGCAGAAGGCTGGGC-3′,3′ primer: 5′-CAAAGTTGTCATGGATGACC-3′.13 To determine the optimal cycles of PCR which represent the quantity of DNA in the linear range, DNA was subjected to 23, 26, 29, and 32 cycles of PCR, and 26 rounds of PCR was chosen (data not shown). The PCR mixture contained 100 ng of DNA, 0.25 pmol/mL of each of the primers, 250 pmol/mL of each of the four deoxyribonucleotide triphosphates (dNTP) (Pharmacia, Stockholm, Sweden), 0.05 U/mL of Taq polymerase (Boehringer Mannheim, Indianapolis, IN) in 20 μL of the manufacturer's specified buffer with 2.0 mmol/L MgCl2 . After 3 minutes of denaturing at 95°C, 26 cycles of denaturing for 40 seconds at 95°C, annealing for 35 seconds at 55°C, and extending for 45 seconds at 72°C were performed in a Programmable Thermal Controller (MJ Research Inc, Watertown, MA). The PCR products were separated on 2% agarose gels and transferred to nylon membranes after staining with ethidum bromide and hybridized with an oligonucleotide probe 5′-TGCAGCAGCTGTTGGTGTACCACAT-3′ for KSHV and 5′-GCAGTTCCAGAGGTCTT-3′ for GAPDH end-labeled with γ-32P dATP using T4 polynucleotide kinase (GIBCO-BRL, Gaithersburg, MD), washed sequentially, and exposed to XOMAT film (Eastman Kodak, Rochester, NY). The beta emissions of each band of the membrane were also assessed by radioanalytic imaging detector Ambis 4000 system (Ambis Inc, San Diego, CA).
Nested PCR.The samples were tested by nested PCR (35 plus 35 cycles) with similar primers used by Monini et al,11 giving a 160-bp band (outer primers 5′-TCCGTGTTGTCTACGTCCAG-3′,5′-AGCCGAAAGGATTCCACCAT-3′; inner primers 5′-ACGGATTTGACCCCGTGTTC-3′,5′-AATGACACATTGGTGGTATA-3′ ). For the first step, the PCR mixture contained 1 μg of DNA. For the ovarian and testicular cancer samples, we used 100 ng of DNA. The DNA was added to 0.25 pmol/mL of each of the outer primers, 250 pmol/mL of each of the four dNTPs, 0.05 U/mL of Taq polymerase in 10 μL of the manufacturer's specified buffer with 2.0 mmol/L MgCl2 . After 3 minutes of denaturing at 95°C, 35 cycles of denaturing for 40 seconds at 95°C, annealing for 35 seconds at 52.5°C, and extending for 45 seconds at 72°C were performed in a Programmable Thermal Controller. For the second step, we added 20 μL of PCR mixture containing 0.25 pmol/mL of each of the inner primers to the 10 μL of PCR product synthesized in the first step. The PCR reaction of the second step was performed in the same manner as the first step. The PCR products were separated on 2% agarose gels and transferred to nylon membranes after ethidum bromide staining and hybridized with an oligonucleotide probe (5′-TGCAGCAGCTGTTGGTGTACCACAT-3′ ) for KSHV which was end-labeled with γ-32P dATP using T4 polynucleotide kinase.
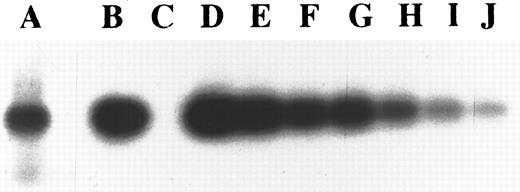
Determination of copy numbers of KSHV in KS-1 cell line. Lane A, 100% KS-1 cell line DNA was amplified with GAPDH primers; lane B, 100% KS-1 cell line DNA; lane C, 100% salmon sperm DNA; lanes D through J, twofold dilutions of KS-1 cell line DNA in salmon sperm DNA, from 1:1 to 1:64. Lanes B through J were amplified with ORF 73 (open reading frame of KSHV) primers. The intensity of the band of GAPDH amplified with 100% KS-1 cell line DNA is close to the intensity of the PCR product of the ORF 73 amplified with a 1:8 dilution of KS-1 cell line DNA. The beta emissions of each band of the membrane were also counted by Ambis 4000; emissions of radioactive lanes A and G were 724 cpm and 750 cpm, respectively.
Determination of copy numbers of KSHV in KS-1 cell line. Lane A, 100% KS-1 cell line DNA was amplified with GAPDH primers; lane B, 100% KS-1 cell line DNA; lane C, 100% salmon sperm DNA; lanes D through J, twofold dilutions of KS-1 cell line DNA in salmon sperm DNA, from 1:1 to 1:64. Lanes B through J were amplified with ORF 73 (open reading frame of KSHV) primers. The intensity of the band of GAPDH amplified with 100% KS-1 cell line DNA is close to the intensity of the PCR product of the ORF 73 amplified with a 1:8 dilution of KS-1 cell line DNA. The beta emissions of each band of the membrane were also counted by Ambis 4000; emissions of radioactive lanes A and G were 724 cpm and 750 cpm, respectively.
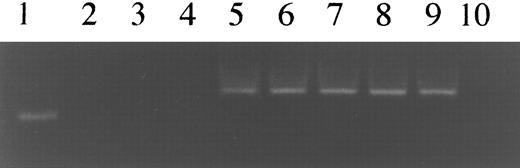
Determination of sensitivity of detection of KSHV sequences. Lane 1, 100-bp DNA ladder; lane 2, K562 cell line DNA; lane 3, HL-60 cell line DNA; lane 4, distilled water. Lanes 2 through 4 served as a negative controls. Lane 5, 10 ng of KS-1 cell line DNA which is estimated to contain 24,000 copies of KSHV sequences. Lanes 5 through 9 are serial 10-fold dilutions of KS-1 cell line DNA in K562 cell line DNA. The 160-bp band that represents the nest-primer product detected up to 2.4 copies of KSHV sequences.
Determination of sensitivity of detection of KSHV sequences. Lane 1, 100-bp DNA ladder; lane 2, K562 cell line DNA; lane 3, HL-60 cell line DNA; lane 4, distilled water. Lanes 2 through 4 served as a negative controls. Lane 5, 10 ng of KS-1 cell line DNA which is estimated to contain 24,000 copies of KSHV sequences. Lanes 5 through 9 are serial 10-fold dilutions of KS-1 cell line DNA in K562 cell line DNA. The 160-bp band that represents the nest-primer product detected up to 2.4 copies of KSHV sequences.
RESULTS
Copy numbers of KSHV in KS-1 cells.To determine the copy numbers of KSHV in KS-1 cells, we made serial twofold dilutions of KS-1 DNA into salmon sperm DNA. The diluted DNA were amplified by 26 cycles of PCR with primers for ORF 73 of KSHV and for GAPDH. The signal of the GAPDH band from undiluted KS-1 cell DNA was compatible with the ORF 73 band from the KS-1 DNA, which was diluted 1:8 (724 counts per minute [cpm] and 750 cpm, respectively, counted by Ambis 4000 System) (Fig 1). Therefore, we estimated that the KS-1 cell line contains about 16 copies of KSHV per cell.
Sensitivity of the nested PCR assay.To determine the sensitivity of our nested PCR assay, the assay was performed on serial 10-fold dilutions of KS-1 DNA, which is estimated to contain 16 copies of KSHV per cell as shown above. The KS-1 DNA was diluted with K562 myeloid leukemia DNA. The nested PCR could detect about 2.4 copies of KSHV sequences on a background of K562 DNA (Fig 2), which means that our assay can detect 1 copy of KSHV in 62,500 KSHV− cells when using 1 μg DNA as template (150,000 cells = 1 μg DNA). No bands were observed in K562 and HL-60 cell lines or distilled water.
Analysis of KSHV sequences in urogenital and prostate tissues.Ten benign prostate specimens from Italian men and 10 from men from the United States, as well as 32 prostatic, 30 vulvar, 20 cervical, 24 ovarian, and 30 testicular cancer specimens from patients from the United States were examined by nested PCR. None of the individuals had a history of HIV infection. None of the samples showed the specific band for KSHV sequences by either ethidum bromide staining or Southern blotting analysis (Fig 3, Table 1).
Nested PCR analysis for KSHV sequences from DNA of urogenital specimens from Italian and American individuals. Lane 1, 100-base pair (bp) molecular weight ladder. Lane 2, 0.01% KS-1 cell line DNA diluted in K562 DNA served as a positive control. The 160-bp band represents the nest-primer product; the two upper bands represent fragments derived from the internal and external primer sets. Lane 3, 100% K562 DNA served as a negative control. Lanes 4 through 13 were DNA samples from normal prostate tissue from Italian men. Lanes 14 through 20 were DNA samples from normal prostate tissue from patients from the United States.
Nested PCR analysis for KSHV sequences from DNA of urogenital specimens from Italian and American individuals. Lane 1, 100-base pair (bp) molecular weight ladder. Lane 2, 0.01% KS-1 cell line DNA diluted in K562 DNA served as a positive control. The 160-bp band represents the nest-primer product; the two upper bands represent fragments derived from the internal and external primer sets. Lane 3, 100% K562 DNA served as a negative control. Lanes 4 through 13 were DNA samples from normal prostate tissue from Italian men. Lanes 14 through 20 were DNA samples from normal prostate tissue from patients from the United States.
KSHV DNA Sequences Analyzed in Urogenital Tissues
| Type of Sample . | No. of Samples . | No. Positive . |
|---|---|---|
| Normal prostate | 20 | 0 |
| Prostate cancer | 32 | 0 |
| Vulvar cancer | 30 | 0 |
| Cervical cancer | 20 | 0 |
| Ovarian cancer | 24 | 0 |
| Testicular cancer | 30 | 0 |
| Type of Sample . | No. of Samples . | No. Positive . |
|---|---|---|
| Normal prostate | 20 | 0 |
| Prostate cancer | 32 | 0 |
| Vulvar cancer | 30 | 0 |
| Cervical cancer | 20 | 0 |
| Ovarian cancer | 24 | 0 |
| Testicular cancer | 30 | 0 |
DISCUSSION
Controversy exists regarding the prevalence of KSHV in the HIV− population.9,10 Recently, Monini et al11 in Italy used nested PCR to detect KSHV sequences in 91% of samples from human sperm, 44% of prostates, 20% of kidneys, 10% of vulva, and 6% of uterine cervical specimens (including carcinomas). They concluded that KSHV infects a large proportion of adults; and like other human herpes viruses, KSHV may be a ubiquitous latent infection in otherwise healthy individuals.11 We studied DNA from 20 normal prostates, as well as 32 prostatic, 30 vulvar, 20 cervical, 24 ovarian, and 30 testicular cancer specimens using similar techniques. In contrast, none of the samples were positive for KSHV sequences.
KS tends to involve certain well-defined ethnic and geographic groups, specifically Central European Jews, Poles, Russians, and Italians.16-18 Because our samples were from both the United States and Italy, the discrepancy between our results and those of Monini et al11 is unlikely to be explained by either ethnic or geographical factors. Therefore, we cannot confirm the findings of Monini et al that KSHV sequences are ubiquitous in urogenital and prostate tissues. Bigoni et al,19 also from Italy, have reported KSHV sequences in 7% to 13% of non-Hodgkin's lymphoma, Hodgkin's disease, reactive lymphadenopathies, and peripheral blood mononuclear cells. They used the same primers that we used in this study and similar nested PCR method (45 cycles plus 45 cycles), but their assay was less sensitive (4 copies per 150,000 cells). One microgram of DNA was used as a template for their first step of nested PCR because they had failed to detect positive results using only 100 ng of DNA as a template, even in samples which showed positive result using 1 μg DNA. They concluded that low virus load might have caused failure of PCR detection of KSHV sequences in previous negative reports.4 20 Because we used 1 μg DNA in most of our samples as a template, our negative results were not caused by low virus loading to the PCR. Therefore, we consider that the samples we studied were indeed negative for KSHV sequences.
The standard nested PCR involves a critical step, passage of the PCR product from the first reaction round of amplification to the second reaction round.21 False positives caused by contamination easily occur using PCR.22 In the United States, Kaposi's sarcoma is at least 20,000 times more common in persons with acquired immunodeficiency syndrome than in the general population and 300 times more common than in other immunosuppressed groups23; thus, KSHV infection is strongly related to immunodeficiency of the host,2,4,24 and with rare exceptions,7 8 KSHV has not been detected in nonimmunosuppressed individuals. Therefore, our results argue against the suggestion that KSHV is a widespread virus in urogenital tissues of healthy individuals.
Supported in part by grants from the National Institutes of Health (DK42792 and CA 42710), the Concern and Cap Cure Foundation, and the Parker Hughes Trust. H.P.K. is a member of the Jonsson Comprehensive Cancer Center and holds the Mark Goodson Chair in Oncology Research.
Address reprint requests to Taizo Tasaka, MD, Cedars Sinai Medical Center, 5th Floor Davis Research Bldg, 8700 Beverly Blvd, Los Angeles, CA 90048.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal