Abstract
We studied the PK-LR gene in 15 unrelated Italian patients with congenital hemolytic anemia associated with erythrocyte pyruvate kinase (PK) deficiency. Fourteen different mutations were detected among 26 mutated alleles identified: a five-nucleotide (nt) deletion (227 to 231), two splice-site (1269C and IVS3(−2)c), 10 missense (514C, 787T, 823A, 993A, 994A, 1168A, 1456T, 1529A, 1552A, and 1594T) and one nonsense mutation(s) (721T). Eight of these (deletion 227-231, 1269C, IVS3(−2)c, 514C, 787T, 823A, 1168A, and 1552A) were novel. Moreover, a new polymorphic site was detected in the 3′ untranslated region of the mRNA (C/T, nucleotide 1738). The deletion 227-231 causes a stop codon after amino acid 77, probably resulting in an unstable gene product. Mutations 1269C and IVS3(−2)c lead to an alteration of the 5′ and 3′ splice-site consensus sequence, respectively; cDNA analysis failed to reveal any abnormal transcript, suggesting that these mutations generate an unstable mRNA that is rapidly degraded. Of the five new missense mutations, 823A (Gly275-Arg) and 1168A (Asp390-Asn) involve highly conserved amino acids, 514C (Glu172-Gln) and 1552A (Arg518-Ser), although found in less conserved regions, affect the balance of the electric charges of the protein. Mutation 787T (Gly263-Trp) is likely to determine strong modifications in the local structure of the molecule. The most frequent mutation in Italy appears to be 1456T (seven of 30 alleles), followed by 1529A (three of 30) and 994A (three of 30). A correlation was found between mutations, biochemical characteristics of the enzyme, and clinical course of the disease.
PYRUVATE KINASE ([PK] adenosine triphosphate [ATP]: pyruvate O2-phosphotransferase, EC 2.7.1.40) is a key enzyme for ATP production in glycolysis. Four isozymes are present in mammalian tissues1-2: L-type (hepatic) and R-type (erythrocytic), encoded by the PK-LR gene under the control of two tissue-specific promoters,3 and M1-(muscle and brain) and M2-(fetal and most adult tissues) types encoded by the PK-M gene by alternative mRNA splicing.4
PK deficiency is the most common cause of hereditary nonspherocytic hemolytic anemia, with nearly 400 cases reported since the first description by Valentine et al5 in 1961. Clinical symptoms usually observed in true homozygotes and compound heterozygotes are variable, ranging from neonatal jaundice requiring exchange transfusions, to a fully compensated hemolytic anemia.6 Many mutant PKs have been identified on the basis of the biochemical characteristics of the defective enzyme.7
The PK-LR gene has been localized on the long arm of chromosome 1,8 and the cDNA of the R-type has been cloned and sequenced: it is 2,060 bp long and codes for 574 amino acids. The codifying region is split into 12 exons, 10 of which are common to the two isoforms, while exons 1 and 2 are specific for the erythrocytic and the hepatic isoenzyme, respectively.9,10
So far, 18 different mutations have been identified in Japanese and Chinese patients10-18 and 57 in caucasians,19-31 with only two mutations (1151T and 1436A) being common to the two populations. Moreover, identical mutations have been associated with different biochemical variants.32
We report the results of a molecular study of 15 Italian patients with PK deficiency. Fourteen different mutations were identified among 26 mutated alleles found — eight of these had not been described previously. Moreover, the molecular results have been related to the biochemical properties of the mutant enzymes and to the clinical pattern.
MATERIALS AND METHODS
Patients. Fifteen unrelated Italian patients with congenital nonspherocytic hemolytic anemia associated with PK deficiency were investigated. The most important clinical and hematologic data at the time of diagnosis are summarized in Table 1. Patients MC, SC, ZM, RG, DM, CA, and SI were newly diagnosed. The clinical history and results of enzyme characterization of the remaining patients have already been reported elsewhere.33-36 In patient DM, ferrokinetic studies showed a sevenfold increase in erythropoiesis associated with peripheral hemolysis and a high degree of ineffective erythropoiesis.37
Clinical and Hematologic Data of PK-Deficient Patients, and the Type of Mutation
| Patient . | Sex . | Origin . | Age (yr) . | Neonatal Jaundice . | ExTx . | Transfusions (U) . | Spleen . | Hb (g/dL) . | Reticulocytes (109/L) . | Unconjugated Bilirubin . | Ferritin (ng/mL) . | TS (%) . | Mutations . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | . | . | . | . | . | (mg/dL) . | . | . | . |
| MM | M | N | 16 | Yes | Yes | 3 | In | 10.4 | 166 | 5.46 | 88 | 33 | ms/ms |
| AI | F | N | 30 | No | No | 0 | In | 11.6 | 167 | 1.27 | 140 | 45 | ms/ms |
| RGC | M | N | 31 | No | No | 0 | In | 12.5 | 123 | 3.57 | 76 | 58 | ms/? |
| MC | F | S | 19 | No | No | 0 | In | 10.0 | 303 | 0.45 | 94 | 43 | ms/? |
| SC | F | S | 2 | Yes | Yes | 0 | In | 11.2 | 169 | 1.03 | 116 | 40 | ms/ms |
| PL | F | N | 43 | No | No | 0 | In | 10.8 | 198 | 1.75 | 186 | 29 | ms/ms |
| DA | F | N | 40 | No | No | 0 | Out | 9.5 | 1,452 | 0.44 | 540 | 90 | ms/ms |
| ZM | M | N | 12 | Yes | Yes | >50 | Out | 8.5 | 263 | 1.59 | 790 | 38 | ms/ms |
| RG | M | S | 6 | Yes | Yes | >50 | Out | 7.4 | 1,046 | 1.97 | 2,800 | 85 | ms/ms |
| IF | M | S | 18 | No | No | 2 | In | 11.6 | 222 | 2.40 | 810 | 30 | ms/ms |
| DM | M | N | 21 | No | Yes | >100 | Out | 7.4 | 1,252 | 5.62 | 716 | 49 | ss/ns |
| CA | F | S | 24 | No | No | 0 | In | 10.6 | 190 | 2.03 | 23 | 15 | ms/ms |
| PP | F | N | 24 | No | No | 0 | In | 12.9 | 130 | 3.97 | 186 | 32 | ms/? |
| FR | F | S | 6 | Yes | Yes | >100 | Out | 8.3 | 113 | 1.56 | 436 | 44 | ss/ms |
| SI | F | S | 6 | Yes | No | >50 | In | 6.9 | 154 | 3.30 | 1,440 | ND | fs/? |
| Reference value | 12.1-16.7 | 24-84 | <0.75 | 19-238 | 16-54 | ||||||||
| Patient . | Sex . | Origin . | Age (yr) . | Neonatal Jaundice . | ExTx . | Transfusions (U) . | Spleen . | Hb (g/dL) . | Reticulocytes (109/L) . | Unconjugated Bilirubin . | Ferritin (ng/mL) . | TS (%) . | Mutations . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | . | . | . | . | . | (mg/dL) . | . | . | . |
| MM | M | N | 16 | Yes | Yes | 3 | In | 10.4 | 166 | 5.46 | 88 | 33 | ms/ms |
| AI | F | N | 30 | No | No | 0 | In | 11.6 | 167 | 1.27 | 140 | 45 | ms/ms |
| RGC | M | N | 31 | No | No | 0 | In | 12.5 | 123 | 3.57 | 76 | 58 | ms/? |
| MC | F | S | 19 | No | No | 0 | In | 10.0 | 303 | 0.45 | 94 | 43 | ms/? |
| SC | F | S | 2 | Yes | Yes | 0 | In | 11.2 | 169 | 1.03 | 116 | 40 | ms/ms |
| PL | F | N | 43 | No | No | 0 | In | 10.8 | 198 | 1.75 | 186 | 29 | ms/ms |
| DA | F | N | 40 | No | No | 0 | Out | 9.5 | 1,452 | 0.44 | 540 | 90 | ms/ms |
| ZM | M | N | 12 | Yes | Yes | >50 | Out | 8.5 | 263 | 1.59 | 790 | 38 | ms/ms |
| RG | M | S | 6 | Yes | Yes | >50 | Out | 7.4 | 1,046 | 1.97 | 2,800 | 85 | ms/ms |
| IF | M | S | 18 | No | No | 2 | In | 11.6 | 222 | 2.40 | 810 | 30 | ms/ms |
| DM | M | N | 21 | No | Yes | >100 | Out | 7.4 | 1,252 | 5.62 | 716 | 49 | ss/ns |
| CA | F | S | 24 | No | No | 0 | In | 10.6 | 190 | 2.03 | 23 | 15 | ms/ms |
| PP | F | N | 24 | No | No | 0 | In | 12.9 | 130 | 3.97 | 186 | 32 | ms/? |
| FR | F | S | 6 | Yes | Yes | >100 | Out | 8.3 | 113 | 1.56 | 436 | 44 | ss/ms |
| SI | F | S | 6 | Yes | No | >50 | In | 6.9 | 154 | 3.30 | 1,440 | ND | fs/? |
| Reference value | 12.1-16.7 | 24-84 | <0.75 | 19-238 | 16-54 | ||||||||
Abbreviations: N, North of Italy; S, South of Italy; ExTx, exchange transfusion; Hb, mean steady-state hemoglobin; TS, transferrin saturation; ND, not determined; ms, missense mutation; ns, nonsense mutation; fs, frameshift; ss, splice site.
The patients underwent the following hematologic investigations: complete blood cell count, reticulocyte count, screening for abnormal or unstable hemoglobin, red blood cell osmotic fragility, autohemolysis, direct antiglobulin test, Ham and sucrose hemolysis test, and assay of the most important erythrocyte enzyme activities of the glycolytic and pentose phosphate pathways. Bilirubin, iron status parameters, transferrin receptor, and erythropoietin concentrations were also determined in the serum. The diagnosis of PK deficiency was made by exclusion of the most common causes of hemolytic anemia and by demonstration of a reduced PK activity in all patients, with the other red blood cell enzymes showing normal or increased activity.
Reference subjects.Fifty normal individuals from various regions in Italy were used as reference population.
Hematologic assays and enzyme studies.Routine hematologic investigations were performed according to the method of Dacie and Lewis.38 The assay of glycolytic and pentose phosphate pathway enzyme activities was performed as reported elsewhere,34 following the methods of Beutler.39 PK was partially purified and characterized according to the International Committee for Standardization in Haematology (ICSH) recommendations.40 Serum iron, total iron-binding capacity, and transferrin saturation were determined by standard methods. Serum ferritin level was measured by an enzyme immunoassay procedure (IMX System, Abbott Laboratories, Abbott Park, IL).
DNA extraction.Blood samples were collected from the patients and normal controls. Leukocytes were isolated and genomic DNA was extracted using standard manual methods.41
Single-strand conformation polymorphism analysis.All 11 exons of the PK-LR gene that code for the R-type isoenzyme were screened by single-strand conformation polymorphism (SSCP) analysis in all patients. Each exon was amplified by polymerase chain reaction (PCR) using 500 ng genomic DNA in a 100-μL mixture containing 33.5 mmol/L Tris hydrochloride, pH 8.8, 1.65 mmol/L MgCl2 , 0.2 mmol/L dNTPs, 100 ng of each primer, and 0.5 U Taq DNA polymerase (Boehringer, Mannheim, Germany) and processed in a Thermalcycler (Perkin-Elmer, Norwalk, CT). The samples were purified with a Qiaex gel extraction kit (Qiagen, Chatsworth, CA) and suspended in 10 μL H2O . Three microliters of sample and 3 μL SSCP loading buffer42 were denatured and loaded on nondenaturing polyacrylamide gel (9% acryl/bis 29:1, 5% glycerol, and 1× TBE). Standard run conditions using a Mini Protean II electrophoretic apparatus (Biorad, Hercules, CA) were as follows: room temperature, 25 V for 30 minutes, 50 V for 30 minutes, 100 V for 1 hour, and 150 V for 2.5 hours. In addition, to increase the sensitivity of SSCP analysis, samples were tested at 4°C with or without glycerol. DNA was stained with silver (Rapid Ag-stain; ICN, Irvine, CA).
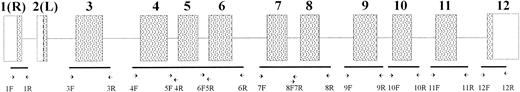
PK-LR gene structure and position of primers used. Boxes show exons: (▧) coding regions: (□) noncoding regions. Exons are indicated by the numbers.
PK-LR gene structure and position of primers used. Boxes show exons: (▧) coding regions: (□) noncoding regions. Exons are indicated by the numbers.
Sequence analysis.SSCP analysis was followed by direct sequencing of the exons in which band shifts were observed. Direct sequencing was made on genomic DNA of the remaining patients until mutations were identified. Amplified DNA was automatically sequenced (A.L.F.; Pharmacia Biotech, Uppsala, Sweden). Direct sequence reactions were performed in solid phase (Dynabeads-280; Dynal, Oslo, Norway); an Auto Read Sequencing kit (Pharmacia Biotech) was used for sequence reactions. Mutations were confirmed by the sequence of both strands.
Oligonucleotides used for SSCP analysis and sequencing were synthesized by a DNA synthesizer (Gene Assembler Plus; Pharmacia Biotech). The sequence and position of the oligonucleotides are summarized in Fig 1 and Table 2.
Oligonucleotides Used for SSCP and Sequence Analysis
| Fragment . | Sense Oligonucleotide . | Antisense Oligonucleotide . | Internal Primers Used for Sequencing . |
|---|---|---|---|
| PK1 | TATTCCATGGTCCCGCAG | GGCTCCTAGTTTTCACCCTC | |
| PK3 | GAGGAAGGGCAGGTGAC | CAAAAGATGAAGAAGCACC | |
| PK4F-6R | GTTGCCTCTCATGTTCTGGG | AATGTGCCCAGCGCACGG | PK4R GGAAGGTGGCCAGGACCTCG |
| PK5F AGGTCCTGGCCACCTTCC | |||
| PK5R CCGCCTTTCCGGCCCTG | |||
| PK6F GACTCCGGGGCTCAGAACT | |||
| PK7F-8R | CAACTGTGCCCCGTCCTCA | CAGGTGTCCCTAAAACCCAC | PK7R GTGATGGGGAATAGCGACAG |
| PK8F CACCTTTCTTCTCCTGCCTG | |||
| PK9 | GTCCTACAACTTTGACATCC | TAGCTCCTCAAACAGCTGC | |
| PK10 | CCTTCCATACCCCAGTGCCC | GCCCACCCCTGACCCAAAGC | |
| PK11 | ACCTTCTCTGCCTCCTCCA | GGGGGGCTCCTGATACAAA | |
| PK12 | TGATTTGTTTTCTTTCCCTC | GTAGGGAGGGTCAGGAAT | |
| PK10-12 cDNA | CACCGCCATTGGTGCTGTGG | GGAAGGGATGGGGTACAAGG |
| Fragment . | Sense Oligonucleotide . | Antisense Oligonucleotide . | Internal Primers Used for Sequencing . |
|---|---|---|---|
| PK1 | TATTCCATGGTCCCGCAG | GGCTCCTAGTTTTCACCCTC | |
| PK3 | GAGGAAGGGCAGGTGAC | CAAAAGATGAAGAAGCACC | |
| PK4F-6R | GTTGCCTCTCATGTTCTGGG | AATGTGCCCAGCGCACGG | PK4R GGAAGGTGGCCAGGACCTCG |
| PK5F AGGTCCTGGCCACCTTCC | |||
| PK5R CCGCCTTTCCGGCCCTG | |||
| PK6F GACTCCGGGGCTCAGAACT | |||
| PK7F-8R | CAACTGTGCCCCGTCCTCA | CAGGTGTCCCTAAAACCCAC | PK7R GTGATGGGGAATAGCGACAG |
| PK8F CACCTTTCTTCTCCTGCCTG | |||
| PK9 | GTCCTACAACTTTGACATCC | TAGCTCCTCAAACAGCTGC | |
| PK10 | CCTTCCATACCCCAGTGCCC | GCCCACCCCTGACCCAAAGC | |
| PK11 | ACCTTCTCTGCCTCCTCCA | GGGGGGCTCCTGATACAAA | |
| PK12 | TGATTTGTTTTCTTTCCCTC | GTAGGGAGGGTCAGGAAT | |
| PK10-12 cDNA | CACCGCCATTGGTGCTGTGG | GGAAGGGATGGGGTACAAGG |
Endonuclease restriction analysis.The oligonucleotides and endonuclease restriction enzymes used (New England Biolabs, Beverly, MA) are reported in Table 3.
Oligonucleotides and Endonuclease Restriction Enzymes Used to Confirm PK Mutations and Polymorphisms
| Mutation . | Exon . | Primers . | Enzyme . | Size of Normal Fragments (bp) . | Size of Mutated Fragments (bp) . |
|---|---|---|---|---|---|
| IVS3(-2)c | IVS3 | PK4F-4R | AvaI | 211 | 157 + 54 |
| 514C | 6 | PK6F-6R | Hinf I | 231 + 51 + 3 | 282 + 3 |
| 721T | 7 | P721-PK7R | Afl III | 411 | 17 + 394 |
| 787T | 7 | PK7F-PK7R | MspI | 122, 108, 104, 41 | 163, 108, 104 |
| 823A | 7 | PK7F-7R | Eco57I | 375 | 201 + 174 |
| 1168A | 9 | PK9F-9R | Bsr DI | 335 | 250 + 85 |
| 1269C | 9 | PK9F-9R | Aci I | 165 + 150 + 20 | 165 + 135 + 35 |
| 1456T | 11 | PK11F-11R | BsmAI | 220 + 36 | 256 |
| 1529A | 11 | PK11F-11R | Sty I | 256 | 144 + 112 |
| 1552A | 11 | PK11-11R | TspRI | 256 | 141 + 115 |
| 1594T | 11 | PK11F-11R | MspI | 175 + 81 | 256 |
| 1705(A/C) | 12 | PK12F-12R | RcaI | 265 (allele A) | 108 + 157 (allele C) |
| 1738(C/T) | 12 | PK12F-12R | BseRI | 265 (allele C) | 130 + 135 (allele T) |
| Mutation . | Exon . | Primers . | Enzyme . | Size of Normal Fragments (bp) . | Size of Mutated Fragments (bp) . |
|---|---|---|---|---|---|
| IVS3(-2)c | IVS3 | PK4F-4R | AvaI | 211 | 157 + 54 |
| 514C | 6 | PK6F-6R | Hinf I | 231 + 51 + 3 | 282 + 3 |
| 721T | 7 | P721-PK7R | Afl III | 411 | 17 + 394 |
| 787T | 7 | PK7F-PK7R | MspI | 122, 108, 104, 41 | 163, 108, 104 |
| 823A | 7 | PK7F-7R | Eco57I | 375 | 201 + 174 |
| 1168A | 9 | PK9F-9R | Bsr DI | 335 | 250 + 85 |
| 1269C | 9 | PK9F-9R | Aci I | 165 + 150 + 20 | 165 + 135 + 35 |
| 1456T | 11 | PK11F-11R | BsmAI | 220 + 36 | 256 |
| 1529A | 11 | PK11F-11R | Sty I | 256 | 144 + 112 |
| 1552A | 11 | PK11-11R | TspRI | 256 | 141 + 115 |
| 1594T | 11 | PK11F-11R | MspI | 175 + 81 | 256 |
| 1705(A/C) | 12 | PK12F-12R | RcaI | 265 (allele A) | 108 + 157 (allele C) |
| 1738(C/T) | 12 | PK12F-12R | BseRI | 265 (allele C) | 130 + 135 (allele T) |
For mutation 721T, which does not create or destroy a restriction endonuclease site, a mismatched oligonucleotide primer (P721) that creates an informative restriction site was used.22 Genomic DNA of 50 normal individuals was investigated for the presence of missense mutations. The PCR was applied to amplify a portion of the PK-LR gene, and restriction endonuclease enzymes were used to identify the presence of mutations.
Reticulocyte mRNA extraction and cDNA analysis.Total RNA was isolated by ammonium chloride lysis43,44 and reverse-transcribed (M-MLV reverse transcriptase; GIBCO BRL, Gaithersburg, MD) to cDNA using random primers.
To study the consequences of mutation IVS3(−2)c for mRNA splicing, cDNA extending from exon 1 to exon 5 (cDNA nucleotide [nt] −39 to nt 468) was amplified by 30 cycles of PCR under the following conditions: 94°C for 30 seconds, 52°C for 30 seconds, and 72°C for 45 seconds. The primers used were sense oligonucleotide 5′-TATTCCATGGTCCCGCAG-3′ and antisense oligonucleotide 5′-GTCCAGGGCGATGGCCAC-3′.
For mutation 1269C, cDNA extending from exon 8 to exon 10 (from nt 1077 to nt 1323) was amplified by 30 cycles of PCR at 94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 45 seconds. The primers used were sense oligonucleotide 5′-CTGCAACTTGGCGGGCAAGC-3′ and antisense oligonucleotide 5′-TAGCTCCTCAAACAGCTGC-3′.
Polymorphism study.Polymorphic sites (nt 1705 and AAT microsatellite in IVS12) were investigated as previously described.13 45 The new polymorphic site at nt 1738 C/T was studied with the restriction endonuclease enzyme Bse RI (New England Biolabs, Beverly, MA) (Table 3).
Computer analysis.Sequences of the new splice-site variant 1269C were obtained using the computer program SPLICE (PCgene; IntelliGenetics, Mountain View, CA).
RESULTS
Table 4 reports molecular and biochemical data in the 15 unrelated PK-deficient patients, together with the respective variant names. Although parental consanguinity was not documented in the families investigated, four patients were found to be homozygous. Fourteen different mutations were detected among 26 mutated alleles identified: deletion 227-231 (TGGAC), two splice-site mutations (nt 1269 GCG-GCC and IVS3(−2)c, one nonsense mutation (nt 721 GAG-TAG), and 10 missense mutations (nt 514 GAG-CAG, 787 GGG-TGG, 823 GGG-AGG, 993 GAC-GAA, 994 GGC-AGC, 1168 GAT-AAT, 1456 CGG-TGG, 1529 CGA-CAA, 1552 CGT-AGT, and 1594 CGG-TGG). Eight mutations (deletion 227-231, 1269C, IVS3(−2)c, 514C, 787T, 823A, 1168A, and 1552A) were novel. Moreover, a new polymorphic site was detected in the 3′ untranslated region of the mRNA (C/T, nt 1738). The frequency of the T allele was similar in patients and controls (28 of 30 v 33 of 36).
Molecular and Biochemical Data of the PK-Deficient Patients
| Patient . | Variant Name . | Mutation . | Amino Acid Substitution . | Activity . | Km PEP (mmol/L) . | Heat Stability4-150 . | Electrophoretic Mobility† . | nt 1705 . | Microsatellite ATT . | nt 1738 . |
|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | (IU/g Hb) . | . | . | . | . | . | . |
| MM | Milano | 1456T/1456T | Arg486-Trp/Arg486-Trp | 4.5 | 1.64 | 66 | 78 | C/C | 14/14 | T/T |
| AI | Soresina | 1456T/1552A | Arg486-Trp/Arg518-Ser | 4.1 | 1.28 | 62 | 86 | C/C | 14/15 | T/T |
| RGC | 1456T/? | Arg486-Trp/? | 3.6 | 4.32 | 53 | 75 | C/C | 14/14 | T/T | |
| MC | 1456T/? | Arg486-Trp/? | 2.8 | 0.84 | 83 | 128 | C/C | 14/14 | T/T | |
| SC | 1456T/823A | Arg486-Trp/Gly275-Arg | 3.0 | 0.35 | 25 | 100 | C/C | 15/19 | T/T | |
| PL | Parma | 1456T/993A | Arg486-Trp/Asp331-Glu | 5.8 | 1.59 | 94 | 100 | C/C | 14/16 | T/T |
| DA | Aprica | 1529A/1529A | Arg510-Gln/Arg510-Gln | 1.4 | 0.78 | 5 | 100 | C/C | 14/14 | T/T |
| ZM | 1529A/994A | Arg510-Gln/Gly332-Ser | 9.7 | 0.96 | 54 | 123 | C/C | 14/14 | T/T | |
| RG | 994A/994A | Gly332-Ser/Gly332-Ser | 3.5 | 0.54 | 29 | 100 | C/C | 17/17 | T/T | |
| IF | Sassari | 514C/514C | Glu172-Gln/Glu172-Gln | 3.1 | 1.32 | 66 | 75 | C/C | 14/14 | T/T |
| DM | IVS3(-2)c/721T | Splice site/Glu241-end | 5.5 | 1.09 | 13 | 88 | C/C | 14/14 | T/T | |
| CA | 1594T/993A | Arg532-Trp/Asp331-Glu | 2.9 | 1.17 | 50 | 120 | C/C | 14/14 | T/T | |
| PP | Mantova | 1168A/? | Asp390-Asn/? | 4.0 | 1.04 | 37 | 87 | C/C | 12/14 | T/T |
| FR | Torre Annunziata | 1269C/787T | Splice site/Gly263-Trp | 2.8 | 13.70 | 78 | 100 | C/C | 13/16 | T/T |
| SI | del 227-231/? | Frame shift/? | 8.3 | 0.17 | 77 | ND | A/A | 12/14 | C/C | |
| Reference value | 11.1-15.59‡ | 0.71-122‡ | 57-99‡ | 100 | ||||||
| Patient . | Variant Name . | Mutation . | Amino Acid Substitution . | Activity . | Km PEP (mmol/L) . | Heat Stability4-150 . | Electrophoretic Mobility† . | nt 1705 . | Microsatellite ATT . | nt 1738 . |
|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | (IU/g Hb) . | . | . | . | . | . | . |
| MM | Milano | 1456T/1456T | Arg486-Trp/Arg486-Trp | 4.5 | 1.64 | 66 | 78 | C/C | 14/14 | T/T |
| AI | Soresina | 1456T/1552A | Arg486-Trp/Arg518-Ser | 4.1 | 1.28 | 62 | 86 | C/C | 14/15 | T/T |
| RGC | 1456T/? | Arg486-Trp/? | 3.6 | 4.32 | 53 | 75 | C/C | 14/14 | T/T | |
| MC | 1456T/? | Arg486-Trp/? | 2.8 | 0.84 | 83 | 128 | C/C | 14/14 | T/T | |
| SC | 1456T/823A | Arg486-Trp/Gly275-Arg | 3.0 | 0.35 | 25 | 100 | C/C | 15/19 | T/T | |
| PL | Parma | 1456T/993A | Arg486-Trp/Asp331-Glu | 5.8 | 1.59 | 94 | 100 | C/C | 14/16 | T/T |
| DA | Aprica | 1529A/1529A | Arg510-Gln/Arg510-Gln | 1.4 | 0.78 | 5 | 100 | C/C | 14/14 | T/T |
| ZM | 1529A/994A | Arg510-Gln/Gly332-Ser | 9.7 | 0.96 | 54 | 123 | C/C | 14/14 | T/T | |
| RG | 994A/994A | Gly332-Ser/Gly332-Ser | 3.5 | 0.54 | 29 | 100 | C/C | 17/17 | T/T | |
| IF | Sassari | 514C/514C | Glu172-Gln/Glu172-Gln | 3.1 | 1.32 | 66 | 75 | C/C | 14/14 | T/T |
| DM | IVS3(-2)c/721T | Splice site/Glu241-end | 5.5 | 1.09 | 13 | 88 | C/C | 14/14 | T/T | |
| CA | 1594T/993A | Arg532-Trp/Asp331-Glu | 2.9 | 1.17 | 50 | 120 | C/C | 14/14 | T/T | |
| PP | Mantova | 1168A/? | Asp390-Asn/? | 4.0 | 1.04 | 37 | 87 | C/C | 12/14 | T/T |
| FR | Torre Annunziata | 1269C/787T | Splice site/Gly263-Trp | 2.8 | 13.70 | 78 | 100 | C/C | 13/16 | T/T |
| SI | del 227-231/? | Frame shift/? | 8.3 | 0.17 | 77 | ND | A/A | 12/14 | C/C | |
| Reference value | 11.1-15.59‡ | 0.71-122‡ | 57-99‡ | 100 | ||||||
Percentage of activity after 60 minutes of incubation at 53°C.
Percentage of normal mobility.
Mean ± 1 SD.
The frame-shift deletion 227-231 causes a stop codon after amino acid 77, probably resulting in an unstable gene product.
Mutation 1269C is localized at the last nt of exon 9 and leads to an alteration of the 5′ splice-site consensus sequence. cDNA analysis of the implicated region failed to reveal any abnormal transcript, and the direct sequence showed only the normal allele. These data suggest that mutation 1269C generates an unstable mRNA that is rapidly degraded.
Mutation IVS3(−2)c (tctcgGGCCA) determines an alteration of the 3′ consensus splice site resulting in an unstable mRNA. This is supported by the finding that no abnormal band was detected at the cDNA level.
None of the five missense mutations were detected in the genomic DNA of 50 normal individuals.
Enzyme characterization of the homozygous patients showed that variants 994A and 1529A were markedly heat-unstable, without alteration of the substrate affinity and electrophoretic pattern, whereas mutants 514C and 1456T displayed abnormal electrophoretic migration.
DISCUSSION
We identified eight new mutations (deletion 227-231, 1269C, IVS3(−2)c, 514C, 787T, 823A, 1168A, and 1552A) in a group of Italian PK-deficient patients. Deletion 227-231 is one of the most drastic PK mutations reported so far. Because the R-type cDNA is 2060 bp long, corresponding to 574 amino acids, this mutation causes a lack of greater than 80% of the normal protein structure. Therefore, the gene product is inactive and unlikely to survive intracellular proteolysis.
Mutations 1269C and IVS3(−2)c involve the consensus sequences for mRNA splicing. The substitution of a single base in this area is known to modify the splicing.46 For mutation 1269C, this is further supported by the observation that another splice-site mutation at the same nt position (1269A) results in PK deficiency.11 Mutation 1269C determines the disruption of the 5′ splice site and activation of a cryptic splice site 18 bases before the normal one, as shown by computer analysis using the Staden method,47 leading to the lack of six amino acid residues. The observation that in our patient cDNA analysis failed to detect any abnormal transcript and that the sequence of this region was normal suggests that the product of allele 1269C is unstable and rapidly degraded. The same conclusion can be drawn for mutation IVS3(−2)c, in which only the normal cDNA was found. In patient DM, this mutation was associated with mutation 721T,21,23 which causes a substitution of Glu 241 to a chain termination codon. According to the crystallographic studies of Muirhead et al,48 the active site of the encoded subunit is not complete, so the gene product is not likely to display PK activity. The combination of two very disruptive mutations may account for the severe clinical and hematologic pattern, characterized by marked peripheral hemolysis associated with a high degree of ineffective erythropoiesis.
For the five new missense mutations, the amino acid changes (Glu172-Gln, Gly263-Trp, Gly275-Arg, Asp390-Asn, and Arg518-Ser) can only be considered the putative disease-producing mutations, since a cause-and-effect relationship between a single amino acid substitution and the enzyme defect is not certain. However, the fact that the rest of the sequence was normal and that none of these mutations were detected in the normal population makes unlikely the possibility of polymorphisms. Moreover, by comparing the amino acid sequences among several species (cat M1, chick M, rat L, yeast, and human),48 we found mutations 823A and 1168A to involve highly conserved amino acids. In particular, the observations that mutation 823A is located close to the putative active site,48 and that a different nt substitution at the same position (823G-C) was associated with hemolytic anemia29 further support the correlation between this mutation and the enzyme defect.
Of the remaining missense mutations, two (514C and 1552A) result in a change of the electric charge of the protein; the former is located in the proximity of the active site region,48 whereas the latter is in a region rich in mutations associated with chronic hemolytic anemia.30 Mutation 787T (Gly263-Trp) is likely to determine strong modifications in the local structure of the molecule.
In this study, we showed that in Italy the most common PK mutation is 1456T (seven of 30 alleles), followed by 1529A (three of 30) and 994A (three of 30); this is different from what was reported in Northern European and US Caucasian populations, where mutation 1529A is the most frequent (42 of 107 alleles) and 1456T is less common (five of 107 alleles).30
Four patients were homozygous for mutations 1456T, 1529A, 994A, and 514C. Although it is known that many factors other than mutant enzyme characteristics may influence the course of the disease,37 information on the clinical pattern of these cases may be of some help in counseling.
The mutation 1456T (patient MM) determines the amino acid substitution Arg486 to Trp and results in slightly increased Km for phosphoenolpyruvate and altered electrophoretic mobility without affecting the enzyme stability. The clinical pattern of this mutation was characterized by neonatal jaundice with a need for exchange transfusion at birth, followed by a lifelong history of moderate anemia with normal iron status.
Mutation 1529A (patient DA) was found in the homozygous state in a large family described elsewhere.34 The mutant enzyme displayed a very low activity and a striking heat instability. This functional abnormality is expected, because Arg 510 is located in the C domains of the protein considered responsible for the interaction between subunits.48 From a clinical point of view, homozygosity for mutation 1529A results in hemolytic anemia that is moderate to severe. Four of five affected siblings required splenectomy and cholecystectomy; of these, two died of overt clinical hemochromatosis and another displayed increased serum ferritin levels in the absence of blood transfusions. This confirms that several factors, including splenectomy, iron overload, and ineffective erythropoiesis, may influence the course of the disease.37
Mutation 994A (patient RG), previously reported by Lenzner et al20 in a compound heterozygous patient, is in a very conserved region near residues directly involved in the active site. The mutation results in enzyme thermal instability and, at the clinical level, in a severe transfusion-dependent hemolytic anemia with unsatisfactory response to splenectomy.
The new mutation 514C, resulting in reduced PK electrophoretic mobility with normal substrate affinity and heat stability, is associated with mild hemolytic anemia and iron overload despite the absence of transfusion support.
Although all the exons and flanking regions were sequenced in all cases, we failed to detect the second mutation in four patients. This is likely due to technical reasons, even though it cannot be excluded that some mutations may be in the regulative regions of the gene or may activate a cryptic splice site in an intron.
As regards the study of polymorphisms, the mutation 1529A was linked with the T allele (nt 1738) and associated with haplotype C, 14 (nt 1705, microsatellite ATT) as reported by others.22 This finding further supports the single origin of this mutation. Mutation 1456 seems to be associated with allele 14 in all cases but one, suggesting either that this mutation arose more than once or that it is a very old mutation. The new polymorphic site 1738, although scarcely informative as a single site, may be useful for family studies when investigated in combination with other markers.
Supported by research Grant No. R.C. 160/01 from IRCCS Ospedale Maggiore of Milan.
Address reprint requests to Alberto Zanella, MD, Divisione di Ematologia, IRCCS Ospedale Maggiore, Via F. Sforza, 35, 20122 Milano, Italy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal