Abstract
Glanzmann thrombasthenia (GT) is a rare bleeding disorder resulting from mutations in either glycoprotein (GP) IIb or GPIIIa genes. The disease is relatively frequent in highly inbred populations such as Iraqi Jews. The molecular basis of GT in 6 unrelated Iraqi-Jewish patients was previously identified as an 11-bp deletion in exon 12 of the GPIIIa gene. We now describe a second mutation found in 3 unrelated Iraqi-Jewish families that consists of an 11.2-kb deletion between an Alu repeat in intron 9 and exon 13 of the GPIIIa gene. The mutant DNA is transcribed into mRNA in which exons 10 through 13 are absent. Splicing of exon 9 directly to exon 14 leads to a shift in the reading frame resulting in a stop codon. The predicted protein is truncated in the middle of the third cysteine-rich domain before the transmembrane domain. Simple DNA-based methods were devised for identification of both mutations in Iraqi Jews for the purpose of carrier detection and prenatal diagnosis enabling prevention of GT. A survey of the general Iraqi-Jewish population for the first 11-bp deletion and the second 11.2-kb deletion disclosed that the allele frequency of the first mutation was 0.0043, whereas none of 700 individuals examined bore the second mutation (allele frequency <0.0007). Among 40 GT patients of Iraqi-Jewish origin 31 were homozygous for the first mutation, 4 were compound heterozygotes for the first and second mutations, and 2 were homozygous for the second mutation. Haplotype analyses using 4 polymorphic markers in the GPIIIa gene showed that each mutation originated in a distinct founder.
THE PLATELET MEMBRANE glycoprotein (GP) complex GPIIb/IIIa (integrin αIIbβ3 ) plays a critical role in platelet aggregation and adhesion due to its capacity to bind fibrinogen, fibronectin, von Willebrand factor, and vitronectin when platelet are activated.1 Glanzmann thrombasthenia (GT) is a severe autosomal recessive bleeding disorder characterized by failure of platelet aggregation in response to adenosine diphosphate (ADP), epinephrine, collagen, and thrombin and by absent or diminished clot retraction.2 These abnormalities are related to severely reduced or dysfunction of the GPIIb/IIIa complex3 that result from mutations in either GPIIb or GPIIIa genes. Most mutations are point mutations (10 in the GPIIIa gene and 12 in the GPIIb gene) and 9 are small or large rearrangements (5 in the GPIIIa gene and 4 in the GPIIb gene).4
GT is relatively frequent in some highly inbred populations such as Indians5; Arabs in Jordan,6 Israel,7 and Saudi Arabia8; Gipsies in France9; and Iraqi Jews residing in Israel.10 The Iraqi-Jewish population has existed as a cohesive community since 586 BC, with intragroup and consanguineous marriage common. A rough estimate of the prevalence of heterozygotes for GT in the general Iraqi-Jewish population disclosed a figure of 1:44.10
The molecular basis of GT in 6 unrelated Iraqi-Jewish patients was identified as an 11-base deletion in exon 12 of the GPIIIa gene.7 In this report, we describe a new mutation found in 3 unrelated Iraqi-Jewish families. This second mutation is also located in the GPIIIa gene and consists of an 11.2-kb deletion between an Alu sequence in intron 9 and exon 13. We show that in the general Iraqi-Jewish population living in Israel the frequency of heterozygotes for the first mutation (I-J1 ) is 1:114 and for the second mutation (I-J2 ) is less than 1:700. Haplotype analyses using 4 polymorphic markers in the GPIIIa gene indicate that each mutation originated in a distinct founder.
MATERIALS AND METHODS
Patients and controls.All patients affected by GT were of Iraqi-Jewish origin. The diagnosis of GT was previously established by history of a life-long mucocutaneous bleeding tendency, a prolonged bleeding time, normal platelet count, absent clot retraction, and failure of platelets to aggregate with ADP, collagen, and epinephrine. Western blot analysis of solubilized platelets performed in most patients showed absent GPIIIa11 and minute amounts of pro-GPIIb.12
The index patient whose examination initiated the present study was a 6-year-old girl who was noted to have facial purpura at birth and has suffered from recurrent spontaneous as well as injury-related bleeding episodes. Her parents are second cousins. A 35-year-old first cousin of the mother is also affected by GT. The parents of the proband wished to have another child and asked to be examined for the purpose of eventual prenatal diagnosis.
Iraqi-Jewish controls were patients consecutively admitted to the Departments of Medicine and Surgery of the Medical Center.13 They were either born in Iraq or had 4 grandparents who were born in Iraq. The study was approved by the Human Ethics Committee of the Sheba Medical Center.
Southern blot analysis.Genomic DNA was prepared from peripheral white blood cells using the protein desalting method.14 The DNA was digested with HindIII restriction enzyme (New England Biolabs Inc, Beverly, MA) and blotted onto Hybond N+ membrane (Amersham, Buckinghamshire, UK). Hybridization was then performed sequentially with labeled fragments of GPIIIa cDNA probes (3.6-kb insert in pGEM-7 [Promega, Madison, WI] digested with Acc I and Not I restriction enzymes [New England Biolabs Inc]) and with five labeled GPIIIa exons obtained by polymerase chain reaction (PCR) that served as genomic probes (see below). All probes were labeled with α-32P-dCTP (Rotem Industries, Beer-Sheva, Israel) using random primer labeling DNA kit (NEBlot; New England Biolabs Inc) according to the manufacturer's instructions.
Platelet mRNA and cDNA preparation.Platelet mRNA was prepared from platelet-rich plasma by affinity chromatography on oligo dT-cellulose (Quick-prep Micro; Pharmacia Biotech, Piscataway, NJ). First-strand cDNA was synthesized by oligo dT primer and avian myeloblastic virus reverse transcriptase (Promega).
Amplification of genomic DNA and cDNA by PCR.The sequences of the oligonucleotides used in this study are detailed in Table 1. Amplification of individual exons of GPIIIa gene was performed with primers flanking them within the respective introns, 15 to 25 bp from the 5′ and 3′ splice junctions.15 Amplification of cDNA was performed with primers located within exons.
Oligonucleotide Primers Used for Amplifications of Genomic DNA or cDNA Fragments
| Location . | Forward (5′ to 3′) . | Reverse (5′ to 3′) . |
|---|---|---|
| Genomic DNA primers* | ||
| Exon 8 | 8F: | 8R: |
| CAGTTCAATTCTTGTCTTCTTGT | GCTCCAGGACAAAGGCCCT | |
| Exon 9 | 9F: | 9R: |
| GGGCCCAACTGTGTCTAAAT | TTATGAGGGGGTGTGGGTT | |
| Exon 10 | 10F: | 10R: |
| CAGCGGGTCCACCTTCCT | CCAGCCTCCCGGTCTCT | |
| Exon 11 | 11F: | 11R: |
| TTGCCTTAATCACTGTGTCCT | CCTGGGTGTGTGCAACTCT | |
| Exon 12 | 12F: | 12R: |
| TTCCTCCATTTTCCCTACTC | CCTGCCTAACATGGTTCTTC | |
| Exon 13 | 13F: | 13R: |
| TTCATTCACAACGCCCTGCT | GACCACCCAAAGCATCTGC | |
| Intron 9† | I9F: | |
| ACTTGGGATCTGAAGTCACAG | ||
| cDNA primers‡ | ||
| Exon 7 | M7F: TTCTGTCCATGGATTCCAGC | |
| Exon 9 | M9F: GTATGCCGTTGTGGGCCTGG | |
| Exon 13 | M13R: TTGCTCTGGCGCGTTCTTCC | |
| Exon 14 | M14R: GTGATATTGGTGAAGGTAGACGTG |
| Location . | Forward (5′ to 3′) . | Reverse (5′ to 3′) . |
|---|---|---|
| Genomic DNA primers* | ||
| Exon 8 | 8F: | 8R: |
| CAGTTCAATTCTTGTCTTCTTGT | GCTCCAGGACAAAGGCCCT | |
| Exon 9 | 9F: | 9R: |
| GGGCCCAACTGTGTCTAAAT | TTATGAGGGGGTGTGGGTT | |
| Exon 10 | 10F: | 10R: |
| CAGCGGGTCCACCTTCCT | CCAGCCTCCCGGTCTCT | |
| Exon 11 | 11F: | 11R: |
| TTGCCTTAATCACTGTGTCCT | CCTGGGTGTGTGCAACTCT | |
| Exon 12 | 12F: | 12R: |
| TTCCTCCATTTTCCCTACTC | CCTGCCTAACATGGTTCTTC | |
| Exon 13 | 13F: | 13R: |
| TTCATTCACAACGCCCTGCT | GACCACCCAAAGCATCTGC | |
| Intron 9† | I9F: | |
| ACTTGGGATCTGAAGTCACAG | ||
| cDNA primers‡ | ||
| Exon 7 | M7F: TTCTGTCCATGGATTCCAGC | |
| Exon 9 | M9F: GTATGCCGTTGTGGGCCTGG | |
| Exon 13 | M13R: TTGCTCTGGCGCGTTCTTCC | |
| Exon 14 | M14R: GTGATATTGGTGAAGGTAGACGTG |
Primers for exons are complementary to intronic sequences flanking each exon.
The primer used is about 100 bp upstream to an Alu repeat.
The primers are contained within exon sequences.
PCRs were performed using different conditions according to the expected length of the DNA products. Products of 150 to 500 bp were amplified by 32 cycles consisting of denaturation for 45 seconds at 94°C, annealing for 45 seconds at 60°C, and extension for 1 minute at 72°C using 0.3 U Taq polymerase (Appligene; Oncor, Pleasanton, CA) in 50 μL volume. Products of 500 to 2,000 bp were amplified by 30 cycles of 1 minute of denaturation at 94°C, 90 seconds of annealing at 60°C, and 3 minutes of extension at 72°C using 1.25 U Taq polymerase (Advanced Biotechnologies, Leatherhead, UK) in 50 μL volume. Products larger than 2,000 bp were amplified using Expand long template PCR system (Boehringer Mannheim Gmbh, Mannheim, Germany) according to the manufacturer's instructions.
Nucleotide sequence analysis.Amplified DNA fragments of 300 to 1,500 bp were recovered from low melt agarose gels using Wizard PCR Preps DNA purification system (Promega). DNA fragments larger than 1,500 bp were purified from agarose gels using Prep-A-Gene DNA purification kit (BioRad, Hercules, CA). Direct sequencing was performed using fmol DNA sequencing system (Promega) and γ-32P–labeled primer according to the manufacturer's instructions. Products were analyzed on 6% polyacrylamide with 7 mol/L urea gels and were subjected to autoradiography.
Survey of the Iraqi-Jewish population for the first and second mutations.The first mutation identified in Iraqi-Jewish patients with GT was searched for in control Iraqi Jews by PCR amplification and agarose gel electrophoresis as described previously.16 The second mutation identified in the present study was examined in the same individuals by PCR amplification and agarose gel electrophoresis as devised during the progress of this work (see Results).
Analysis of polymorphisms in the GPIIIa gene.Four polymorphic sites within the GPIIIa gene were analyzed in genomic DNA of GT patients, their family members, and 120 randomly selected Iraqi-Jewish controls. These were the dimorphic human platelet alloantigen 1 (HPA-1) in exon 3 detected by PCR amplification and Msp I digestion17; a multiallelic polymorphism of CT repeats in intron 6 detected by PCR amplification of the corresponding DNA region in the presence of α-32P-dCTP (Rotem Industries) and analysis in denaturing polyacrylamide gel electrophoresis and autoradiography18; a dimorphism involving substitution of A by C in nucleotide 1163 in exon 8 creating a Taq I cleavage site; and a dimorphism involving substitution of A by G in nucleotide 1565 in exon 9 that eliminates a Sma I restriction site.19 The latter two dimorphisms were detected by PCR amplification using primers flanking the respective exon and were digested with the respective enzyme.
RESULTS
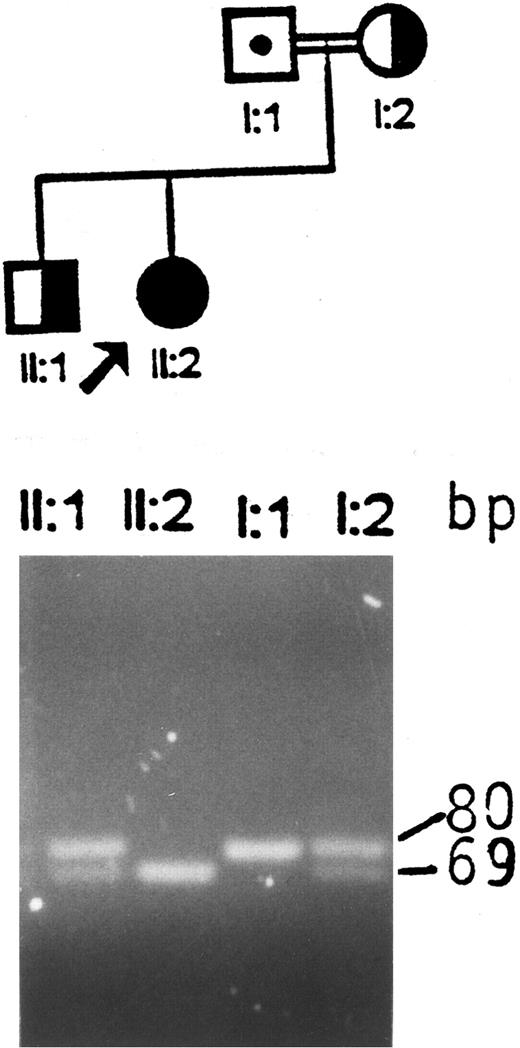
Identification of an aberrant HindIII fragment in the GPIIIa gene of the index patient and her father.Screening for the first Iraqi-Jewish mutation (I-J1 ) in the GPIIIa gene in the index family disclosed that the proband was an apparent homozygote, the mother and a sibling were heterozygotes, and the father appeared normal (Fig 1). The possibilities of uniparental isodisomy (nondisjunction of chromosome 17) and nonpaternity of the father were excluded by analyses of microsatelite markers (data not shown). Germinal mosaicism was also excluded by obtaining identical results in analyses of the I-J1 mutation in the father's sperm cells and white blood cells.
Analysis of the I-J1 mutation in the proband and her immediate relatives. An apparent 11-bp deletion is seen in the PCR product of the proband, ie, a 69-bp fragment instead of the normal 80-bp fragment. The mother and the brother are heterozygous and have both 69- and 80-bp fragments. The father, who is an obligate carrier, shows only the 80-bp fragment, which is consistent with a normal genotype.
Analysis of the I-J1 mutation in the proband and her immediate relatives. An apparent 11-bp deletion is seen in the PCR product of the proband, ie, a 69-bp fragment instead of the normal 80-bp fragment. The mother and the brother are heterozygous and have both 69- and 80-bp fragments. The father, who is an obligate carrier, shows only the 80-bp fragment, which is consistent with a normal genotype.
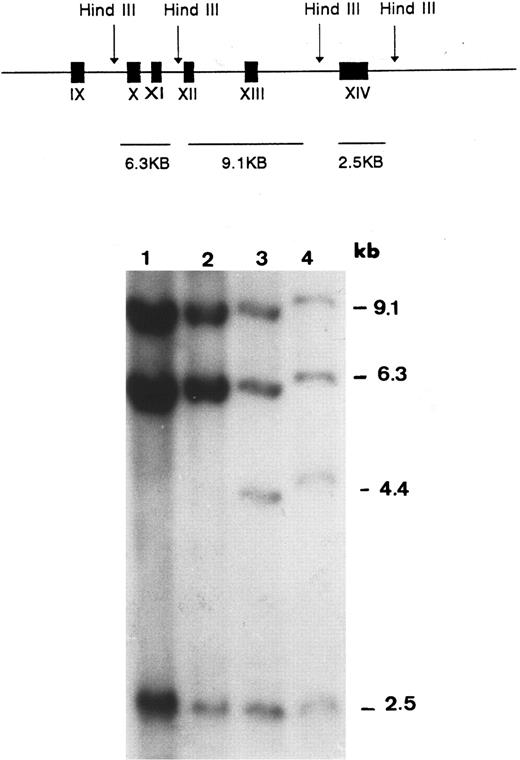
The possibility of a null allele due to a major gene aberration was then examined by Southern blot hybridization. Using Acc I-Not I fragment encompassing exons 10 through 14 of the GPIIIa cDNA as a probe, three fragments of HindIII-digested genomic DNA (9.1, 6.3, and 2.5 kb) were demonstrable in a control and the mother (lanes 1 and 2 in Fig 2). In the genomic DNA of the father (lane 3) and the proband (lane 4), an additional band of 4.4 kb was observed. Rehybridization with probes of individual exons 10, 11, 12, 13, and 14 showed that the 4.4-kb aberrant fragment observed in the proband and her father contained only exon 13. Hybridization with Not I-Not I fragment of GPIIIa cDNA encompassing exons 1 through 9 showed an identical pattern in the proband, her father, and the control DNA (data not shown). The father was also found to be heterozygous for the Sma I polymorphism in exon 9, indicating that this exon was present in the two copies of his chromosomes. These findings were consistent with a large deletion flanked by exons 9 and 13.
Southern blot analysis of HindIII-digested genomic DNA probed with GPIIIa cDNA encompassing exons 10 through 14. The expected HindIII fragments in normal genomic DNA are graphically illustrated in the upper panel. In the autoradiogram, an extra band (4.4 kb) is seen in DNA samples of the proband (lane 4) and her father (lane 3). This band is absent in the sample of the mother (lane 2) and in the sample of a normal control (lane 1).
Southern blot analysis of HindIII-digested genomic DNA probed with GPIIIa cDNA encompassing exons 10 through 14. The expected HindIII fragments in normal genomic DNA are graphically illustrated in the upper panel. In the autoradiogram, an extra band (4.4 kb) is seen in DNA samples of the proband (lane 4) and her father (lane 3). This band is absent in the sample of the mother (lane 2) and in the sample of a normal control (lane 1).
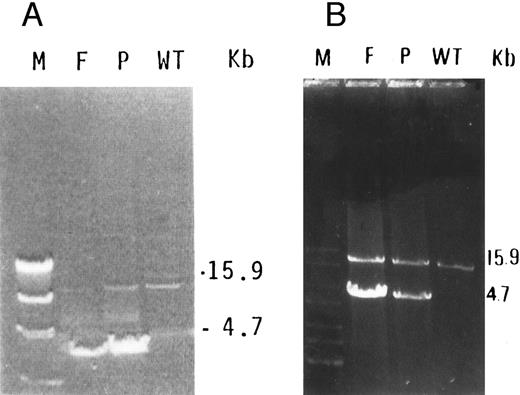
Characterization of a large deletion in the mutant genomic DNA.We predicted that, by using a forward primer upstream to exon 9 (9F ) and a reverse primer downstream to exon 13 (13R) in a long PCR system, the amplification of mutant DNA would yield a smaller fragment than the control DNA. Indeed, a 15.9-kb fragment was obtained in normal DNA, whereas, in the proband and her father, an additional fragment of 4.7 kb was observed (Fig 3). Because the normal fragment of 15.9 kb was very faint in the DNA of the father, it was reamplified using the same forward primer and a reverse primer derived from exon 13 at its 3′ end (M13R, Table 1). This yielded clear 15.9-kb bands in the father, proband, and normal DNA (Fig 3B).
(A) Long PCR of genomic DNA using primers flanking exons 9 (9F ) and 13 (13R) of the GPIIIa gene. A normally amplified 15.9-kb fragment is seen in control DNA (WT) in the proband (P) and very faintly in her father (F ). An additional 4.7-kb fragment is seen in both the proband and her father. The λ DNA molecular weight marker cleaved with HindIII (M) is shown in the left lane. (B) Reamplification of the previous PCR products using reverse primer in exon 13 (M13R) and the same forward primer (9F ) shows more clearly the 15.9-kb fragment in the normal allele, in the proband (P), and in her father (F ).
(A) Long PCR of genomic DNA using primers flanking exons 9 (9F ) and 13 (13R) of the GPIIIa gene. A normally amplified 15.9-kb fragment is seen in control DNA (WT) in the proband (P) and very faintly in her father (F ). An additional 4.7-kb fragment is seen in both the proband and her father. The λ DNA molecular weight marker cleaved with HindIII (M) is shown in the left lane. (B) Reamplification of the previous PCR products using reverse primer in exon 13 (M13R) and the same forward primer (9F ) shows more clearly the 15.9-kb fragment in the normal allele, in the proband (P), and in her father (F ).
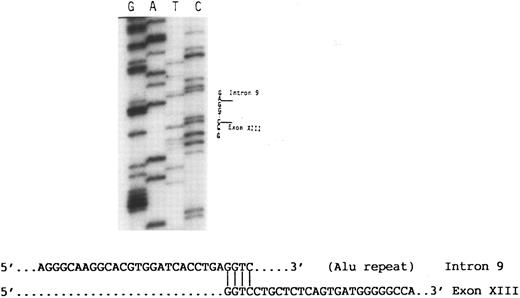
The 4.7-kb fragment of the proband was then recovered from the agarose gel and used as template for further PCR amplifications of exons 9, 10, 11, 12, and 13 using the appropriate primers (Table 1). These experiments showed that only exon 9 was amplified, indicating that exons 10, 11, and 12 and at least part of exon 13 were absent from this fragment (data not shown). Using a primer 3′ to exon 13 (13R) in nucleotide sequence analysis of the 4.7-kb fragment disclosed a sequence consisting of the last 139 nucleotides of exon 13 followed by a sequence derived from an Alu repeat in intron 9 (Fig 4). At the junction between intron 9 and exon 13 there was a GGTC sequence. This sequence is present in both intron 9 and exon 13 of normal DNA, and thus the breakpoint that created the large deletion could have occurred before the G, between G and G, G and T, T and C, or after C. Based on the published sequence of the GPIIIa gene,19 the data indicate that the I-J2 mutation is an 11.2-kb deletion.
Partial nucleotide sequence analysis of the abnormal 4.7-kb fragment (Fig 3). A sequence of an Alu repeat in intron 9 is followed by a sequence derived from exon 13. Note that the homologous nucleotide sequence GGTC in intron 9 and exon 13 at their junction appears only once in the abnormal fragment.
Partial nucleotide sequence analysis of the abnormal 4.7-kb fragment (Fig 3). A sequence of an Alu repeat in intron 9 is followed by a sequence derived from exon 13. Note that the homologous nucleotide sequence GGTC in intron 9 and exon 13 at their junction appears only once in the abnormal fragment.
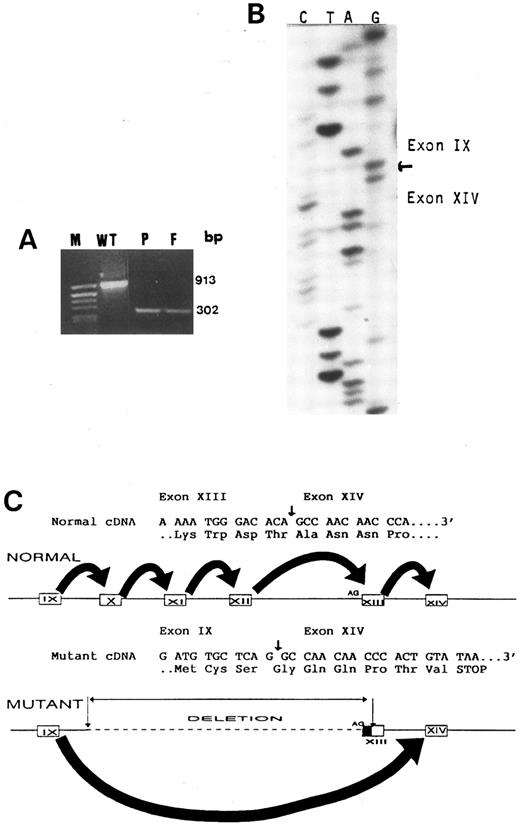
Mutant mRNA consists of exon 9 directly spliced to exon 14.Reverse transcribed cDNA prepared from platelet mRNA of the proband, her father, and a normal control was subjected to PCR using a forward primer in exon 9 and a reverse primer in exon 14 (M9F and M14R in Table 1). The amplified products for exons 9 through 14 were 913 bp in control cDNA (as expected) and 302 bp in the cDNA of the proband and her father (Fig 5). This finding was consistent with a deletion of 611 bp in the mutant cDNA due to skipping of exons 10, 11, 12, and 13. The 913-bp fragment were not demonstrable in the father and proband (Fig 5), probably due to preferential amplification of the mutant cDNA, which was one third in size of normal cDNA.20 Splicing of exon 9 directly to exon 14 in the mutant cDNA was confirmed by sequencing of the 302-bp fragment that was recovered from the agarose gel. Figure 5 shows that, in normal and mutant cDNA, exon 9 ends with G, which is the first nucleotide of a Gly codon (GGC). Because normal exon 14 starts with a GCC codon for Ala, the observed direct splicing of exon 9 to 14 in the mutant cDNA is predicted to cause a shift in the reading frame leading to translation of 6 abberant amino acids followed by a stop codon.
(A) PCR amplification of platelet cDNA (RT-mRNA) using primers flanking exons 9 and 14 shows a 913-bp fragment in a control (WT) and a 302-bp fragment in the proband (P) and her father (F ). The molecular weight marker pBR322 DNA cleaved with Alu I (M) is in the left lane. (B) Nucleotide sequence analysis of the mutant cDNA showing that exon 9 is followed by exon 14. (C) A schematic representation of both normal and mutant genes and their respective mRNA splicings. Because the large deletion includes the AG acceptor site of exon 13, exon 9 is spliced directly to exon 14, skipping the remaining part of exon 13. Direct splicing of exon 9 to 14 in the mutant cDNA is predicted to cause a shift in the reading frame, creating a stop codon after 6 abberant amino acids.
(A) PCR amplification of platelet cDNA (RT-mRNA) using primers flanking exons 9 and 14 shows a 913-bp fragment in a control (WT) and a 302-bp fragment in the proband (P) and her father (F ). The molecular weight marker pBR322 DNA cleaved with Alu I (M) is in the left lane. (B) Nucleotide sequence analysis of the mutant cDNA showing that exon 9 is followed by exon 14. (C) A schematic representation of both normal and mutant genes and their respective mRNA splicings. Because the large deletion includes the AG acceptor site of exon 13, exon 9 is spliced directly to exon 14, skipping the remaining part of exon 13. Direct splicing of exon 9 to 14 in the mutant cDNA is predicted to cause a shift in the reading frame, creating a stop codon after 6 abberant amino acids.
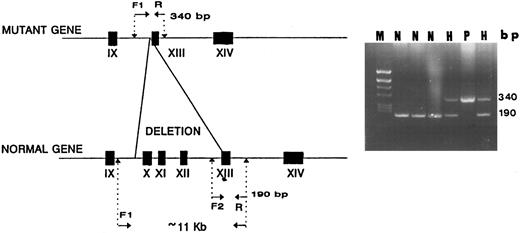
Design of a method for simple detection of the second Iraqi-Jewish mutation.Primers flanking the observed 11.2-kb deletion were synthesized. The forward primer had a sequence derived from intron 9 upstream to the Alu repeat (I9F in Table 1) and the reverse primer consisted of a sequence in exon 13 downstream to the deletion (M13R in Table 1). PCR with these primers yielded a 340-bp fragment only in mutant genomic DNA; in normal DNA, no amplification took place under the condition used because of the excessive size of the predicted fragment (>11.2 kb). Using an alternative forward primer (13F in Table 1) that is derived from a sequence 5′ to exon 13, ie, in the deletion, the PCR yielded an 190-bp fragment only in control DNA and not in mutant DNA (Fig 6). Using all 3 primers (I9F, M13R, and 13F in Table 1) in the same PCR reaction yielded both 190- and 340-bp fragments in the proband and her father.
A diagnostic PCR simultaneously using 3 primers for detection of the I-J2 mutation. F1 and R primers yield a 340-bp fragment only in DNA of a chromosome bearing the mutation as in a homozygous patient (P). In normal DNA (N), no amplification occurs due to the large distance between the primers (<11 kb). F2 and R primers yield a 190-bp fragment only in normal DNA, with no amplification occurring in mutant DNA because F2 is located within the deletion (P). In two hetrozygotes (H), both 190- and 340-bp fragments are demonstrable.
A diagnostic PCR simultaneously using 3 primers for detection of the I-J2 mutation. F1 and R primers yield a 340-bp fragment only in DNA of a chromosome bearing the mutation as in a homozygous patient (P). In normal DNA (N), no amplification occurs due to the large distance between the primers (<11 kb). F2 and R primers yield a 190-bp fragment only in normal DNA, with no amplification occurring in mutant DNA because F2 is located within the deletion (P). In two hetrozygotes (H), both 190- and 340-bp fragments are demonstrable.
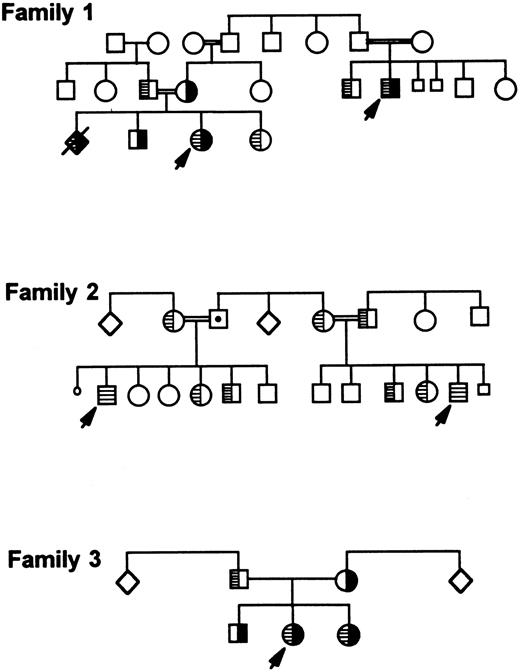
Detection of the second mutation in additional families.Screening of most Iraqi-Jewish GT patients and their family members living in Israel for the second mutation (I-J2 ) showed two additional unrelated families whose members carried it (Fig 7, families 2 and 3). In one of the families (family 2), two cousins were homozygous for the I-J2 mutation. In the second family (family 3), two siblings were compound heterozygotes for the I-J1 and the I-J2 mutations. Their father was heterozygous for the I-J2 mutation and the mother was heterozygous for the I-J1 mutation. In the index family (family 1), the additional affected relative and an affected fetus (diagnosed prenatally and aborted) were compound heterozygotes for both mutations I-J1 and I-J2 (Fig 7). Extensive questioning of several elderly persons belonging to each of the 3 families failed to disclose any relationship among them.
Three Iraqi-Jewish families carrying both I-J1 and I-J2 mutations or only the I-J2 mutation. I-J1 mutation is shown in black, and I-J2 mutation is marked by hatching inside symbols. Family 1 is the index family that depicts an aborted fetus who was prenatally diagnosed as a compound heterozygote similarly to the proband.
Three Iraqi-Jewish families carrying both I-J1 and I-J2 mutations or only the I-J2 mutation. I-J1 mutation is shown in black, and I-J2 mutation is marked by hatching inside symbols. Family 1 is the index family that depicts an aborted fetus who was prenatally diagnosed as a compound heterozygote similarly to the proband.
The distribution of the 2 mutations among Iraqi-Jewish GT patients in Israel is summarized in Table 2. Thirty-one patients are homozygotes for the I-J1 mutation, 4 are compound heterozygotes for both mutations, and 2 are homozygotes for the I-J2 mutation. Of the remaining patients, 2 siblings probably bear a third mutation, because neither I-J1 nor I-J2 mutations were detectable and one was lost from the follow-up.
Distribution of the Mutations Causing GT in Iraqi-Jewish Patients
| Genotype* . | Patients . | Families . |
|---|---|---|
| I-J1/I-J1 | 31† | 16 |
| I-J2/I-J2 | 2 | 1 |
| I-J1/I-J2 | 4 | 2 |
| Undefined‡ | 2 | 1 |
| Not examinedρ | 1 | 1 |
| Total | 40 | 21 |
| Genotype* . | Patients . | Families . |
|---|---|---|
| I-J1/I-J1 | 31† | 16 |
| I-J2/I-J2 | 2 | 1 |
| I-J1/I-J2 | 4 | 2 |
| Undefined‡ | 2 | 1 |
| Not examinedρ | 1 | 1 |
| Total | 40 | 21 |
I-J1 is an 11-bp deletion in exon 12. I-J2 is an 11.2-kb deletion of exons 10 through 13 (see text).
In five of the patients, the genotype was inferred from the genotypes of their immediate relatives.
Two siblings that bear neither I-J1 nor I-J2 mutations.
ρ A patient lost from follow-up.
Prevalence of the I-J1 and I-J2 mutations in the general Iraqi-Jewish population.Of 700 Iraqi-Jewish controls, 6 were found to be heterozygous for I-J1 mutation and none was found to bear the I-J2 mutation. Thus, the frequency of the I-J1 mutant allele in this population is 0.0043 (95% confidence interval, 0.0016 to 0.0093) and the frequency of the I-J2 mutant allele is less than 0.0007 (95% confidence interval, 0 to 0.0026).
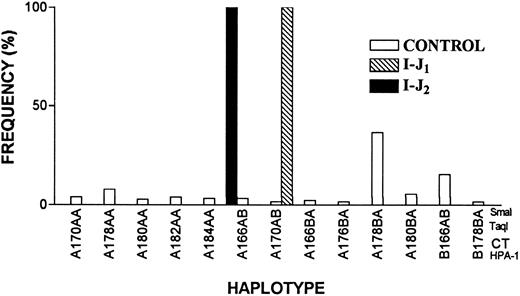
Distinct founder effects for I-J1 and I-J2 mutations.A possible founder effect for both mutations or distinct founder effects for each mutation were examined by haplotype analyses. Using 4 polymorphic markers, we analyzed 62 chromosomes bearing the I-J1 mutation, 8 chromosomes bearing the I-J2 mutation, and 240 chromosomes of control Iraqi Jews (Table 3). Figure 8 shows that all chromosomes carrying the I-J1 mutation had the same haplotype of the GPIIIa gene, which was very rare in the controls (1.7%). All chromosomes bearing the I-J2 mutation had a haplotype that was distinct from the I-J1 haplotype and was also rare (3.4%) in controls (based on 174 informative alleles of control Iraqi Jews).
Allele Frequencies (%) of Polymorphic Markers in the GPIIIa Gene in Iraqi-Jewish Controls and GT Patients Bearing I-J1 and I-J2 Mutations
| Polymorphism . | Allele3-150 . | Control . | Patients Bearing . | |
|---|---|---|---|---|
| . | . | (240)3-151 . | Mutation . | |
| . | . | . | I-J1 (62)3-151 . | I-J2 (8)3-151 . |
| HPA-1 | A | 84 | 100 | 100 |
| B | 16 | 0 | 0 | |
| CT repeat | 166 | 25 | 0 | 100 |
| 170 | 3 | 100 | 0 | |
| 174 | 1 | 0 | 0 | |
| 176 | 5 | 0 | 0 | |
| 178 | 40 | 0 | 0 | |
| 180 | 14 | 0 | 0 | |
| 182 | 10 | 0 | 0 | |
| 184 | 2 | 0 | 0 | |
| Taq I | A | 53 | 100 | 100 |
| B | 47 | 0 | 0 | |
| Sma I | A | 74 | 0 | 0 |
| B | 26 | 100 | 100 | |
| Polymorphism . | Allele3-150 . | Control . | Patients Bearing . | |
|---|---|---|---|---|
| . | . | (240)3-151 . | Mutation . | |
| . | . | . | I-J1 (62)3-151 . | I-J2 (8)3-151 . |
| HPA-1 | A | 84 | 100 | 100 |
| B | 16 | 0 | 0 | |
| CT repeat | 166 | 25 | 0 | 100 |
| 170 | 3 | 100 | 0 | |
| 174 | 1 | 0 | 0 | |
| 176 | 5 | 0 | 0 | |
| 178 | 40 | 0 | 0 | |
| 180 | 14 | 0 | 0 | |
| 182 | 10 | 0 | 0 | |
| 184 | 2 | 0 | 0 | |
| Taq I | A | 53 | 100 | 100 |
| B | 47 | 0 | 0 | |
| Sma I | A | 74 | 0 | 0 |
| B | 26 | 100 | 100 | |
A designates the digested fragment. B designates the undigested fragment. Numbers indicate the size (in basepairs) of the amplified genomic DNA fragments.
The numbers in parentheses indicate the number of alleles analyzed.
Frequency distribution of GPIIIa haplotypes in 174 informative chromosomes of control Iraqi Jews and 70 chromosomes of GT patients. Note that the haplotypes of 62 chromosomes bearing I-J1 mutation and 8 chromosomes bearing I-J2 mutation are different and that each of them is very rare in the general population.
Frequency distribution of GPIIIa haplotypes in 174 informative chromosomes of control Iraqi Jews and 70 chromosomes of GT patients. Note that the haplotypes of 62 chromosomes bearing I-J1 mutation and 8 chromosomes bearing I-J2 mutation are different and that each of them is very rare in the general population.
DISCUSSION
In a previous study, we defined the molecular basis of GT in 6 of 21 families belonging to the Iraqi-Jewish community as an 11-bp deletion in exon 12 of GPIIIa gene.7 Because this cohesive population is relatively small, with 270,000 of its members living in Israel,10 and because the community has been isolated for about 2,500 years with a high rate of inbreeding, our early finding inferred that all Iraqi-Jewish GT patients carry the same mutation. The present study shows that this assumption was wrong. The second mutation defined herein in 3 unrelated Iraqi-Jewish families is a large 11.2-kb deletion in the GPIIIa gene. The rearrangement consists of joining an Alu repetitive element in intron 9 with a sequence within exon 13. The resultant skipping of the AG acceptor site of exon 13 induces a forced splicing between exon 9 and exon 14 that leads to a frame shift and stop codon. The predicted truncated GPIIIa terminates in the middle of the third cysteine-rich domain before the transmembrane domain. By immunoblotting we were unable to show any GPIIIa in platelets of either a homozygote for the I-J2 mutation or a compound heterozygote for the I-J1 and the I-J2 mutations (data not shown). This finding is similar to the one obtained in homozygotes for the I-J1 mutation11 also consisting of a stop codon before the transmembrane domain.7 Notably, the severity of bleeding manifestations in homozygotes for the I-J1 mutation, homozygotes for the I-J2 mutation, and compound heterozygotes for the I-J1 and I-J2 mutations were indistinguishable.
The presence of a deletion has been suggested sometimes to increase the likelihood of the evolvement of additional events.21 We therefore addressed the question of whether one mutation may have lead to the occurrence of the other particularly, because both are located at similar sites and present in a confined population. The possibility that I-J1 mutation stemmed from I-J2 mutation is refutable, because the site of the former is inside the large deletion of the I-J2 mutation. An opposite sequence of events, ie, that the I-J2 mutation stemmed from the I-J1 mutation, is also unlikely in view of the haplotype analyses (Fig 8) that indicated that each mutation occurred on a distinct rare chromosome.
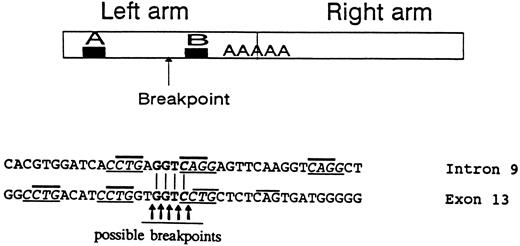
Large gene deletions were reported to result from illegitimate recombinations involving Alu sequences.22,23 The GPIIIa gene may be particularly prone to such events because it possesses 1 Alu per 2 kb,19 whereas the human genome contains 1 Alu per 5 kb on average.24 Indeed, a large inversion-deletion involving 3 Alu sequences was reported in the GPIIIa gene,25 and the present work describes a second large rearrangement involving an Alu repeat. The 5′ breakpoint of the deletion in intron 9 occurred in the left arm of the Alu repeat between 2 sequences homologous to promoters of genes transcribed by RNA polymerase III designated A and B in Fig 9.26 Eighty-nine percent of breakpoints within Alu repeats occur in this region,22 indicating its predisposition to breakage possibly by the conformational changes occurring during transcription. The joining sequence of intron 9 and exon 13 was found to contain GGTC. This 4-nucleotide sequence is present in both intron 9 and exon 13 (Fig 9); consequently, the breakpoint could have occurred at 5 sites, either before, inside, or after the GGTC sequence. The 3′ breakpoint of the I-J2 mutation in exon 13 is in a non-Alu sequence. Previous studies of large rearrangements have shown that, adjacent to breakpoints in Alu and non-Alu sequences, there are usually 4 to 7 direct or inverted nucleotide repeats.27,28 Such repeats were observed in intron 9 and exon 13 close to the breakpoint as follows: four CCTG repeats (3 in exon 13 and 1 in intron 9) as well as 2 complementary CAGG sequences in intron 9 (Fig 9). Moreover, CTG or complementary CAG repeats were demonstrable very close to the possible breakpoints in intron 9 and exon 13. Such CTG/CAG repeats have been suggested to be recognition sites for topoisomerase I potentially involved in illegitimate recombinations and rearrangements.29 It is conceivabe, therefore, that the I-J2 mutation in the founder evolved by an illegitimate recombination that occured when topoisomerase and/or RNA polymerase III were bound to single-stranded DNA.
The upper panel represents a typical Alu repeat consisting of left and right arms with the poly A linker. The A and B regions are sequences homologous to promoters of genes transcribed by RNA polymerase III. The arrow indicates the presumed 5′ breakpoint in intron 9 of the GPIIIa gene creating the I-J2 mutation. The lower panel shows the sequences in proximity to the breakpoints in intron 9 and in exon 13. The GGTC identical sequence in intron 9 and exon 13 is in bold letters, with the putative breakpoints depicted. Direct/inverted repeats frequently observed close to breakpoints are underlined. CTG/CAG repeats suggested to be recognition sites for topoisomerase I are marked by bold lines above letters.
The upper panel represents a typical Alu repeat consisting of left and right arms with the poly A linker. The A and B regions are sequences homologous to promoters of genes transcribed by RNA polymerase III. The arrow indicates the presumed 5′ breakpoint in intron 9 of the GPIIIa gene creating the I-J2 mutation. The lower panel shows the sequences in proximity to the breakpoints in intron 9 and in exon 13. The GGTC identical sequence in intron 9 and exon 13 is in bold letters, with the putative breakpoints depicted. Direct/inverted repeats frequently observed close to breakpoints are underlined. CTG/CAG repeats suggested to be recognition sites for topoisomerase I are marked by bold lines above letters.
During the last 25 years, the diagnosis of GT was made in 40 patients belonging to 21 unrelated Iraqi-Jewish families living in Israel (Table 2). Twenty of the 40 patients were descendants of consanguineous marriages. Our previous study7 and the present work have defined the mutant genotypes in 37 of the patients. Table 2 shows that the predominant mutation is I-J1 , which was found in 66 of the 80 mutant chromosomes. I-J2 mutation was found in only 8 of the 80 mutant chromosomes. Of the 6 remaining mutant chromosomes, 4 (in 2 siblings) bear neither I-J1 nor I-J2 mutations and are currently being investigated and one patient was lost from our follow-up.
In the general Iraqi-Jewish population, heterozygosity for the I-J1 mutation was detected in 6 of 700 individuals, ie, an allele frequency of 0.0043. None of these individuals carried the I-J2 mutation; thus, the exact frequency of this allele is at present unknown but is apparently less than 0.0007. The proportion of more than 6:1 between the frequencies of I-J1 versus I-J2 alleles in the general Iraqi-Jewish population is similar to the 8:1 proportion of mutant chromosomes in thrombasthenic patients (66 v 8 chromosomes). These figures allow us to roughly estimate the risk of births of new thrombasthenic patients in the general Iraqi-Jewish population. If the frequency of heterozygotes for the I-J1 mutation is 10:1,140 and for the I-J2 mutation perhaps 1:1,140, this would yield a 1:100 heterozygosity (11:1,140) rate for both mutations in this population. Consequently, there is a 1:10,000 chance for unrelated individuals who get married to be both affected by either mutations and a 1:40,000 chance of having an affected offspring. These chances would be of course significantly higher if one or both parents are related to one of the affected families.
During the last 25 years we have noticed that patients affected by GT due to the I-J1 and/or I-J2 mutations are significantly handicapped by their life-long moderate to severe bleeding manifestations, particularly epistaxis, gingival bleeding, and menorrhagia. Most families have chosen as a consequence not to have children after the birth of an affected child. The described PCR-based methods for detection of the I-J1 and I-J2 mutations have provided us with reliable means of carrier detection and prenatal diagnosis. At the time of this writing, we have performed 11 prenatal diagnoses during the 10th week of gestation by chorionic villus sampling in 5 women who had previously delivered an affected child. Four of the fetuses were homozygous for the I-J1 mutation and one was a compound heterozygote for both mutations; consequently, all were aborted at the wish of the parents. Four fetuses were heterozygotes for the I-J1 mutation, one was heterozygous for the I-J2 mutation, and one had a normal genotype. All of these pregnancies already had reached term, with none of the infants presenting with bleeding manifestations.
ACKNOWLEDGMENT
We are indebted to Shoshana Yakar and Sally Usher for their skillful technical assistance and to Dr Ilya Novikov for his help with statistical analyses.
Supported by the Israel Science Foundation administered by the Israel Academy of Sciences and Humanities.
Address reprint requests to Uri Seligsohn, MD, Institute of Thrombosis and Hemostasis, Department of Hematology, Chaim Sheba Medical Center, Tel-Hashomer 52621, Israel.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal