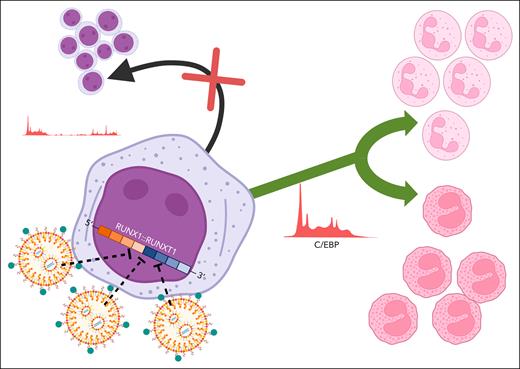
In this issue of Blood, Derevyanko et al1 use an innovative lipid nanoparticle (LNP) delivery system and chemically modified small interfering RNAs (siRNAs) to target the RUNX1::RUNX1T1 fusion breakpoint in samples from patients with acute myeloid leukemia (AML). They find that transient silencing of RUNX1::RUNX1T1 fusion gene expression is sufficient to decrease stem cell self-renewal and cause partial myeloid differentiation toward either a neutrophil or eosinophil cell fate (see figure).
Delivery of siRNA complementary to the fusion site of RUNX1::RUNX1T1 messenger RNA into primary human AML cells is improved nearly 10-fold with the use of targeted lipid nanoparticles. Transient silencing of RUNX1::RUNX1T1 disrupts leukemia stem cell renewal (black arrow) and is associated with increased expression of C/EBP and bidirectional myeloid differentiation into neutrophils or eosinophils (green arrow). Figure created with BioRender.com. Obeng E. (2025) https://BioRender.com/jfqkgjz.
Delivery of siRNA complementary to the fusion site of RUNX1::RUNX1T1 messenger RNA into primary human AML cells is improved nearly 10-fold with the use of targeted lipid nanoparticles. Transient silencing of RUNX1::RUNX1T1 disrupts leukemia stem cell renewal (black arrow) and is associated with increased expression of C/EBP and bidirectional myeloid differentiation into neutrophils or eosinophils (green arrow). Figure created with BioRender.com. Obeng E. (2025) https://BioRender.com/jfqkgjz.
Chromosomal abnormalities are present in ∼50% of patients with AML. The reciprocal translocation between chromosomes 8 and 21, t(8;21)(q22:q22), is most prevalent in children and young adults (12%-15% of cases) and results in the generation of the RUNX1::RUNX1T1 fusion transcript.2 Although t(8:21) AML is generally associated with favorable risk disease, ∼50% of patients with RUNX1::RUNX1T1 AML will fail to achieve long term disease free survival after treatment with cytotoxic chemotherapy due to drug resistance or relapse.3,4 Targeted therapies that induce a more durable remission with less toxicity are needed to improve outcomes for these patients.
The t(8;21) translocation causes a preleukemic condition associated with increased stem cell self-renewal and a block in myeloid differentiation.4 The transcription factor RUNX1 is a master regulator of hematopoietic stem cell development and differentiation. RUNX1T1 is a transcriptional corepressor that recruits histone deacetylase complexes to suppress gene expression. The RUNX1::RUNX1T1 fusion protein causes transcriptional and epigenetic dysregulation by competing with full length RUNX1 for binding to DNA via the N-terminal Runt-homology domain and then silencing the expression of hematopoietic genes and tumor suppressors by recruiting repressive complexes with the RUNX1T1 C-terminal domain.5
Derevyanko et al build on previous studies of a modified siRNA that is complementary to and specific for the RUNX1::RUNX1T1 fusion site. This siRNA has been shown to prolong the survival of immunodeficient mice transplanted with a human t(8;21) AML cell line and decrease leukemia engraftment in secondary transplant recipients.6 As primary human AML cells are notoriously difficult to transduce,7 the authors improved the function of the LNP siRNA delivery system by incorporating a ligand that targets very late antigen-4, an integrin expressed on hematopoietic stem and progenitor cells and mature blood cells.1 This modification in the LNPs led to a 9-fold improvement in siRNA uptake by patient samples after 24 hours. Transient exposure of patient samples to an siRNA targeting the RUNX1::RUNX1T1 fusion site led to a 3-fold decrease in fusion protein expression that persisted for over 10 days. This decrease in RUNX1::RUNX1T1 protein expression was sufficient to block engraftment of a patient-derived xenograft (PDX) cell line into immunodeficient mice (0% compared to 60% engraftment of PDX cells exposed to control siRNA). Similar to the findings that incorporation of all-trans retinoic acid (ATRA) and arsenic trioxide into the treatment of patients with acute promyelocytic leukemia transformed a highly fatal AML subtype into one with a very favorable prognosis due to the induction of terminal differentiation of leukemic blasts,8 Derevyanko et al also demonstrate that siRNA targeting of the RUNX1::RUNX1T1 fusion site leads to bidirectional differentiation of t(8:21) patient samples ex vivo. Although these findings suggest that RUNX1::RUNX1T1 fusion-targeting siRNA may be a feasible strategy to improve treatment efficacy and decrease the incidence of late effects caused by exposure to conventional chemotherapy, the strong effect of the fusion-targeting siRNA on engraftment and the inability to trace primary patient material after xenotransplantation make it difficult to assess whether the bidirectional differentiation observed ex vivo in high cytokine media also occurred in vivo after xenotransplantation.
Mechanistically, the authors interrogated gene expression and chromatin accessibility in patient samples treated with control or fusion-targeting siRNA and found that knockdown of RUNX1:RUNX1T1 messenger RNA was associated with decreased chromatin accessibility in hematopoietic stem and progenitor cells, newly accessible chromatin regions in monocytes and neutrophils, and increased expression of CCAAT/enhancer-binding protein (C/EBP) genes. Integrated analyses of single cell RNA sequencing data generated from patient samples treated with control or fusion-targeting siRNA and publicly available single cell RNA sequencing data suggest t(8;21) AML cells are capable of undergoing limited differentiation toward neutrophil or eosinophil cell fates after RUNX1:RUNX1T1 knockdown and this may be related to the upregulation of different C/EBP family members.1 Future studies evaluating whether targeted knockdown of CEBPA can prevent myeloid differentiation compared to how enforced expression of CEBPE or CEBPD influences eosinophil and/or neutrophil maturation in t(8;21) AML cells exposed to fusion-targeting siRNA will provide further mechanistic insights into how the expression of different C/EBP family members alters myeloid differentiation in cells with the RUNX1::RUNX1T1 gene fusion.
This study provides important insights into the role of RUNX1::RUNX1T1 expression in leukemia stem cell maintenance and moves siRNA therapy closer to clinical application. Transcription factors such as RUNX1 are notoriously difficult to target due to the presence of intrinsically disordered regions and the lack of “druggable pockets” for small molecules to bind.9 Efficient targeting of gene fusions with fusion-specific siRNAs may provide a potent and selective therapy with significantly less toxicity than conventional chemotherapy for the treatment of AML. In addition to this critical advance in the delivery of an siRNA targeting a transcription factor fusion to primary patient samples, the field is also exploring alternative transcription factor-targeting approaches including molecular degraders and proteolysis targeting chimeras.10
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal