In this issue of Blood, Parker et al1 demonstrate the use of methylation sequencing technology to monitor clonal dynamics in 34 patients with clonal hematopoiesis (CH) from the Vanderbilt University Clonal Hematopoiesis and Inflammation in the Vasculature (CHIVE) cohort.
Clinical surveillance of patients with CH involves tracking variant allele fraction (VAF) and complete blood counts (CBCs) for evidence of clonal and hematologic progression, respectively.2 Interpretation of serial VAF and CBC measurements must account for clinical contexts that influence cell type proportions, including infection, inflammatory disease, or exposure to cytotoxic therapies. Parker et al used a targeted enzymatic DNA methylation sequencing assay that captures nearly 4 million 5′-C-phosphate-G-3′ sites (CpGs), combined with methylCC, a previously described statistical method3 to deconvolute cellular composition and classify the clonal behavior in sample pairs as “stagnant,” “expanding,” or “shrinking.” Because of the linear relationship between changes in VAF and cell-type proportions, classifications of clonal behavior derived from DNA methylation-predicted cell-type proportions were more accurate than those based solely on measured changes in VAF (see figure).
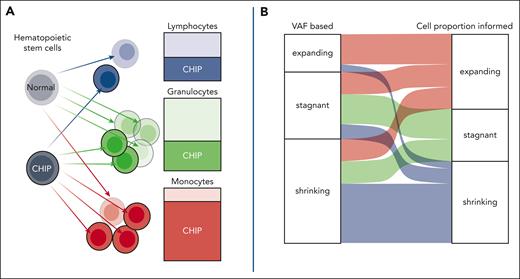
Driver mutations in CH are found in distinct proportions of lymphocytes, granulocytes, and monocytes. (A) The cellular composition of blood samples significantly influences VAF measurements; thus, changes in VAF are susceptible to confounding by exposure and clinical conditions influencing cell type proportions. (B) Cell proportion informed predictions of VAF using data from methylation sequencing result in more accurate predictions of clonal trajectories, reclassifying the behavior of 57.1% of clones. CHIP, clonal hematopoiesis of indeterminate potential. See Figures 1A and 2D in the article by Parker et al that begins on page 988.
Driver mutations in CH are found in distinct proportions of lymphocytes, granulocytes, and monocytes. (A) The cellular composition of blood samples significantly influences VAF measurements; thus, changes in VAF are susceptible to confounding by exposure and clinical conditions influencing cell type proportions. (B) Cell proportion informed predictions of VAF using data from methylation sequencing result in more accurate predictions of clonal trajectories, reclassifying the behavior of 57.1% of clones. CHIP, clonal hematopoiesis of indeterminate potential. See Figures 1A and 2D in the article by Parker et al that begins on page 988.
Genotype-specific variation in cellular proportions occurs with distinct skewing of myelopoiesis depending on the driver gene mutation. For instance, TET2 mutant clones bias toward increased proinflammatory monocyte production.4,5 Consistent with this, Parker et al showed that predictions of clonal behavior are driver gene specific, with distinct equations governing predicted changes in VAF for TET2-mutant, DNMT3A-mutant, and other gene-mutant CH. Estimated VAF changes for TET2-mutant CH relied heavily on increased monocyte proportions, whereas estimates of DNMT3A-clonal behavior relied on decreasing monocytes. Although the authors benchmarked the performance of their methodology in a separate cohort of 106 patients from Vanderbilt’s BioVU biobank, larger-scale data sets will be required to examine the full extent to which this method accurately predicts driver gene- and variant-specific differences in clonal behavior.
Though clonal dynamics may have relevance in predicting clinical trajectories, recent large-scale efforts to derive tools to estimate the risk of myeloid neoplasia in CH have relied on single time point genomic data annotated with longitudinal clinical outcomes.6,7 Resulting models, such as the clonal hematopoiesis risk score (CHRS)6 allow for the identification of individuals with CH at particularly high risk for malignant progression, facilitating risk-specific management in specialized hematology clinics and participant selection for therapeutic clinical trial participation.2 Driver gene-specific differences in clonal growth rates as previously demonstrated in data sets with serial VAF measurements8,9 may have a strong influence on clinical outcomes in CH. Moreover, context-dependent differences in clinical trajectories are perhaps best exemplified by the observation that TP53 mutations are not strongly associated with progression to myeloid malignancy in healthy populations with age-related CH,6 whereas expansion and selection of TP53-mutant clones increase the risk of therapy-related myeloid neoplasms in chemotherapy-exposed solid tumor populations10 As such, the accuracy of risk prediction in the CHRS and other risk prediction models may be substantially improved by data that inform the clinical context in which CH is detected and clonal dynamics (kinetics and direction of change in clone size).
In the future, multimodal analysis of serial samples from prospective cohorts, such as CHIVE, may allow the combined interpretation of VAF and cell type proportions within the context of well-annotated clinical and environmental exposures to more accurately estimate clinical trajectories in CH. Although we await the maturity and widespread availability of such cohorts, reliable next VAF predictions from DNA methylation sequencing data by Parker et al suggest that the addition of DNA methylation assays into the routine assessment of CH may improve the accuracy of risk predictions from single time point data.
Conflict-of-interest disclosure: L.D.W. declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal