In this issue of Blood, Laurent et al dissect the heterogeneity of follicular lymphoma (FL) and present a new clinically applicable model that captures the biological diversity of the disease.1 Several prognostic models, based either on gene expression or genetic features, have been proposed for FL.2-4 Although these can potentially aid in risk stratification for patients treated with rituximab-chemotherapy (R-chemo), they can be difficult to implement in routine practice and are rarely portable to other regimens. Also, although the available models can help identify patients with higher risk of progression or histologic transformation, none offer guidance for a specific treatment when deviating from standard of care. This represents a significant knowledge gap in the face of a growing number of promising treatment options.
The first genetic prognostic model for FL that gained popularity is M7-FLIPI. Here, mutations in EZH2, MEF2B, or ARID1A each predict a superior outcome and mutations in any of EP300, CARD11, CREBBP, and FOXO1 predict inferior outcomes.3 Appealing for ease of implementation, M7-FLIPI provides a prognostic score derived from clinical and genetic features rather than grouping patients with unified biological features. Using mutations from a larger set of genes, Crouch et al used an unsupervised approach to divide FLs into 3 groups with distinct patterns of driver mutations and noncoding mutations.5 Contrasting the genetics of FL to de novo diffuse large B-cell lymphoma (DLBCL), Dreval et al proposed two genetic subgroups: constrained (cFL) and DLBCL-like (dFL).6 Although this established a framework for identifying FLs with a higher risk of transformation (ie, cFL), none of the genetic groups in either study was associated with progression-free survival (PFS). The work hinted at the existence of distinct subgroups within FL and yet genetics alone seemed insufficient to fully capture their biological differences.
The utility of gene expression profiling for prognostication has been firmly established in diffuse DLBCL, particularly with the cell-of-origin (COO) and dark zone signature (DZsig) subgroups.7 The existence of a comparable COO structure for FL that explains the clinical heterogeneity has been proposed but the molecular features have remained elusive. Using gene and protein expression (immunohistochemistry), Mottok et al demonstrated that FL biopsies had variable FOXP1 expression.8 They also noted that mutations in some genes, such as EZH2 and MEF2B, were more common in FOXP1-low tumors. FOXP1 positivity was strongly associated with inferior outcomes for FL patients treated with R-chemo. Using a supervised analysis, Huet and colleagues derived a signature associated with shorter PFS in FL and found this gene set (ICA13) was enriched for genes expressed in centroblasts.4 When the prognostic association of this signature was confirmed by another team, it was found to be highly correlated with FOXP1 expression.2 A more recent study using single-cell gene and protein profiling also proposed two distinct categories of FL.9 In this case, tumor cells in the first category resembled germinal center B cells (GC), whereas the second had molecular features of pre–class switch recombination, post-GC memory B cells. Collectively, the gene expression studies alluded to the existence of at least 2 FL subgroups while leaving the cell-of-origin question unresolved.
In their study, Laurent et al approached the problem by applying both genetic and gene expression profiling to biopsies from patients treated upfront with either R-chemo or rituximab-lenalidomide (R2) in the RELEVANCE trial. Starting with the gene expression data, they identified sets of genes with coordinated expression across the tumors. Two gene sets (CC17 and CC21) were anticorrelated, with some tumors exhibiting high expression of CC17 genes and others expressing CC21 genes. By comparing these to single-cell gene expression profiles derived from healthy tonsil, they attributed CC17 to GC B cells and CC21 to memory B cells. Reasoning that these signatures reflect the cell of origin, they named the FLs expressing CC17 GC-like and the others memory (MEM)-like. To allow reproducible assignment of biopsies to either category, they implemented a 20-gene model (FL20) that divides patients into these groups. Recognizing the need for an approach that can be applied clinically, they also developed a 4-gene immunohistochemistry algorithm that was highly concordant with FL20 assignments. Using both methods, patients with MEM-like FL had inferior PFS on the R-chemo arm relative to those with GC-like FL. Strikingly, although the association with poor prognosis was confirmed in additional external R-chemo cohorts, the inferior outcomes were not experienced by the MEM-like FL patients treated with R2.
The authors have formalized an approach to subdivide FL that bridges the gaps between the proposed genetic models and gene expression signatures. FL20 is highly correlated with the gene expression signatures from the earlier studies.2,4,8 In each case, the high-risk group exhibited high expression of TCF4 or FOXP1---both defining features of MEM-like FL (see figure). Intriguingly, 4 of the M7-FLIPI genes had mutations enriched in one or the other FL20 subgroup, suggesting that the prognostic utility of that model is grounded, in part, in COO. The mutations enriched in MEM-like FL are nearly identical to the defining genetic features of the cFL observed by Dreval et al (see figure), but the overlap with dFL is less clear. Importantly, dFL was defined mostly by noncoding mutations, many outside the regions queried by exome sequencing. Thus, although they are beginning to converge, the relationship between the genetic and gene expression subgroups will require more thorough exploration.
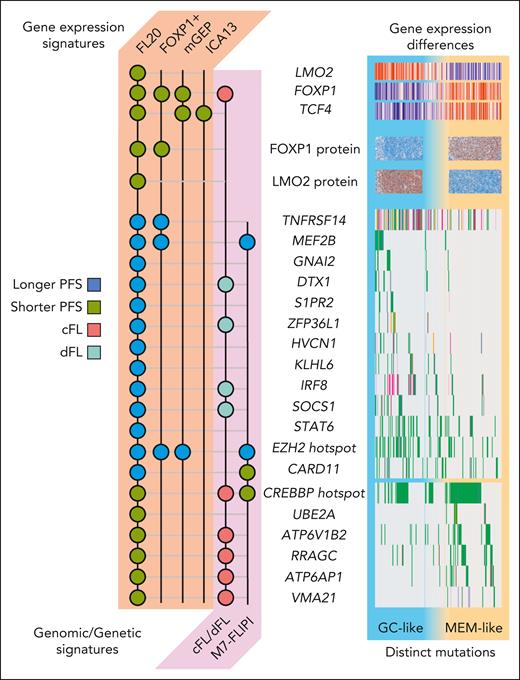
Molecular features of the new FL COO subgroups. The mutation and gene expression features shared with FL20 and previous FL models/subgroups are summarized. Each circle indicates a gene/feature that is associated with the patient category or survival correlate indicated in the legend. The models based on gene or protein expression are FOXP1 positivity from Mottok et al,8 the mGEP signature from Silva et al,2 and the ICA13 signature from Huet et al.4 The models based on FL genetics are the M7-FLIPI from Pastore et al3 and the genomic model described by Dreval et al.6 The right-side portion of the illustration summarizes these features across the cohort analyzed by Laurent et al. Patients are arranged with those assigned to the GC-like subgroup on the left and the MEM-like patients on the right. Only genes reported by Laurent et al to be mutated more in GC-like or MEM-like FL are shown.
Molecular features of the new FL COO subgroups. The mutation and gene expression features shared with FL20 and previous FL models/subgroups are summarized. Each circle indicates a gene/feature that is associated with the patient category or survival correlate indicated in the legend. The models based on gene or protein expression are FOXP1 positivity from Mottok et al,8 the mGEP signature from Silva et al,2 and the ICA13 signature from Huet et al.4 The models based on FL genetics are the M7-FLIPI from Pastore et al3 and the genomic model described by Dreval et al.6 The right-side portion of the illustration summarizes these features across the cohort analyzed by Laurent et al. Patients are arranged with those assigned to the GC-like subgroup on the left and the MEM-like patients on the right. Only genes reported by Laurent et al to be mutated more in GC-like or MEM-like FL are shown.
This new COO-based model of FL appears prognostic for R-chemo and may allow for identification of patients who could benefit from R2 in the frontline setting. Paramount next steps include testing the reproducibility of the immunohistochemistry algorithm in other clinical labs and, ideally, adaptation to a gene expression assay that can be applied clinically. The prognostic utility of this model must also be evaluated in patients treated with other frontline regimens, particularly those in common use, such as bendamustine-rituximab, and in the relapse setting. Finally, we cannot overlook the possibility that unique molecular features of MEM-like FL may represent targetable vulnerabilities. Indeed, several mutations imply their reliance on autophagy, which could theoretically be targeted with existing therapies that are not in use for FL.10
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal