Key Points
Phospho-serine RUNX1 increases upon megakaryocytic fate specification, and phosphomimetic mutant RUNX1 promotes megakaryocytic fate in MEP.
CDK9-induced phosphorylation of RUNX1 in MEPs promotes megakaryocytic fate at the expense of erythroid fate.
Visual Abstract
The specification of megakaryocytic (Mk) or erythroid (E) lineages from primary human megakaryocytic-erythroid progenitors (MEPs) is crucial for hematopoietic homeostasis, yet the underlying mechanisms regulating fate specification remain elusive. In this study, we identify RUNX1 as a key modulator of gene expression during MEP fate specification. Overexpression of RUNX1 in primary human MEPs promotes Mk specification, whereas pan-RUNX inhibition favors E specification. Although total RUNX1 levels do not differ between Mk progenitors (MkPs) and E progenitors (ErPs), there are higher levels of serine-phosphorylated RUNX1 in MkPs than ErPs, and mutant RUNX1 with phosphorylated-serine/threonine mimetic mutations (RUNX1-4D) significantly enhances the functional efficacy of RUNX1. To model the effects of RUNX1 variants, we use human erythroleukemia (HEL) cell lines expressing wild-type (WT), phosphomimetic (RUNX1-4D), and nonphosphorylatable (RUNX1-4A) mutants showing that the 3 forms of RUNX1 differentially regulate expression of 2625 genes. Both WT and RUNX1-4D variants increase expression in 40%, and decrease expression in another 40%, with lesser effects of RUNX1-4A. We find a significant overlap between the upregulated genes in WT and RUNX1-4D–expressing HEL cells and those upregulated in primary human MkPs vs MEPs. Although inhibition of known RUNX1 serine/threonine kinases does not affect phosphoserine RUNX1 levels in primary MEPs, specific inhibition of cyclin dependent kinase 9 (CDK9) in MEPs leads to both decreased RUNX1 phosphorylation and increased E commitment. Collectively, our findings show that serine/threonine phosphorylation of RUNX1 promotes Mk fate specification and introduce a novel kinase for RUNX1 linking the fundamental transcriptional machinery with activation of a cell type–specific transcription factor.
Introduction
Human megakaryocytic-erythroid progenitors (MEPs) give rise to megakaryocytic progenitors (MkPs) or erythroid progenitors (ErPs). Although there may be alternate routes to Mk commitment,1 multiple studies, including our own, demonstrate the presence of functionally bipotent MEPs.2 Using highly enriched primary adult human MEPs,3 single-cell sequencing and subsequent use of chemical (all-trans retinoic acid or rapamycin) and molecular approaches revealed that the cell cycle intrinsically affects fate with fast-cycling biasing MEPs toward the E fate and slower cycling biasing MEPs toward the Mk fate.4,5 We have also reported that iron availability affects MEP fate specification, with low iron promoting megakaryocytic fate specification at the expense of erythroid.6 Thus, MEP fate specification is regulated by both intrinsic and extrinsic factors.
The transcription factor RUNX1 (also called acute myeloid leukemia 1 [AML1] or core binding factor α [CBFα]), 1 of the most commonly mutated genes in acute myeloid leukemia (AML), has been extensively studied in hematopoiesis. RUNX1 and its homologues (RUNX2 and RUNX3) have Runt homology DNA binding domains and require heterodimerization with CBFβ, which stabilizes RUNX1 DNA binding for transcriptional activity.7 RUNX1 can act as transcriptional activator or suppressor.8 In adult mice, inducible knock out of Runx1 decreases megakaryocyte number, Mk maturation, and platelet count.9 Patients with inherited loss-of-function RUNX1 mutations have familial platelet disorder with a high risk of AML.10 Although RUNX1 inhibits E and promotes Mk differentiation of primary murine and human hematopoietic stem and progenitor cells,11,12 whether RUNX1 plays a role in fate specification at the level of the MEP has not previously been determined.
Here, we show that RUNX1 phosphorylated on serine and threonine (S/T) residues promotes Mk and decreases E specification of primary human MEP. We found that a phosphomimetic mutant of RUNX1 was more effective in inducing Mk fate in MEPs compared with wild-type (WT) or mutant RUNX1 that prevents phosphorylation of specific S/T residues. Molecular studies of WT and mutant forms of RUNX1 in human erythroid leukemia (HEL) cells revealed that the phosphomimetic RUNX1 mutant was more effective in inducing gene expression changes in HEL cells with upregulation and downregulation of Mk-associated and E-associated genes, respectively, despite minimal differences in DNA association. Gene expression changes closely paralleled the differentially expressed genes (DEGs) in primary human MEPs undergoing fate specification. We identified cyclin dependent kinase 9 (CDK9) as a novel kinase that phosphorylates RUNX1 in MEPs and showed that CDK9 downregulation in MEPs results in decreased RUNX1 phosphorylation and increased erythroid specification.
Materials and methods
Cell source and cell sorting
Cells from healthy granulocyte–colony stimulating factor–mobilized donors were purchased commercially (AllCells, Alameda, CA), and CD34 selected by the Yale Cooperative Center of Excellence in Hematology. Common myeloid progenitors (CMPs), MEPs, ErPs, and MkPs were sorted on a BD FACSAria, as previously described.3 HEL cells were purchased from the American Type Culture Collection.
Viral transduction
MEPs were cultured in expansion media, as previously described,3 for 16 hours and transduced in the presence of polybrene (8 μg/mL); 24 hours after transduction, cells were counted and plated for colony forming unit (CFU) assays. HEL cells were transduced with virus and 8 μg/mL polybrene and selected in 2 μg/mL puromycin. Murine stem cell virus–driven retroviral plasmid constructs for hemagglutinin (HA)-tagged RUNX1-4A, RUNX1-WT, and RUNX1-4D13 were provided by Dong-Er Zhang (University of California, San Diego, CA).
Drug treatment
Ro5-3335 (Calbiochem) and AI-14-9114 (provided by John H. Bushweller, University of Virginia) were dissolved in dimethyl sulfoxide (DMSO). MEPs were cultured in expansion media3 for 8 hours before drug treatment. Drugs were added to the indicated final concentrations for the indicated times before plating cells for CFU assay in the absence of drugs.
MegaCult-based CFU assays
Transduced or drug-treated MEPs were plated in MegaCult C Medium plus Lipids (STEMCELL Technologies) as previously described.6 Transduced MEPs were selected with 0.5 μg/mL puromycin in MegaCult. After 2 weeks, colonies were stained overnight with fluorophore-conjugated lineage-specific antibodies (CD235 for E and CD41 for Mk) added for a final 1:300 dilution in the semisolid MegaCult media. Colonies were scored based on fluorescent staining.6
Intracellular staining and flow cytometry
Cells were fixed and permeabilized with Lyse/Fix Buffer and Perm Buffer II (BD Biosciences) according to the manufacturer’s protocols. Primary antibodies (phosphorylated serine 276 [pS276]-RUNX1, pS303-RUNX1, or total RUNX1) were used at 1:100 dilution, and secondary antibodies at 1:50 dilution were added at room temperature for 30 minutes, and cells analyzed using a BD LSR II flow cytometer and FlowJo software. Primary antibody targeting pS303-RUNX115 was provided by Alan Friedman (Johns Hopkins University). Additional details in supplemental Material, available on the Blood website.
CUT&RUN and RNA sequencing
HEL cells (500 000 cells per sample) were used for CUT&RUN assays16 performed (Epicypher CUTANA) following the manufacturer’s instructions. DNA library preparation and sequencing were performed by the Yale Center for Genomic Analysis. RNA was prepared using a RNAqueous-Micro kit (Invitrogen) per manufacturer’s instructions. DNA libraries were prepared using TruSeq Stranded messenger RNA kit (Illumina). Data analysis details are provided in supplemental Materials.
Statistical analyses
To account for donor- and batch-specific effects in each experiment, paired t tests were used to assess statistical significance for most assays. For high-throughput data, adjusted P values were used to account for multiple comparison testing.
Results
RUNX1 promotes megakaryocytic fate in human MEP
Analysis of our published single-cell RNA-sequencing (scRNA-seq) data5 indicates that DEGs between CMPs, MEPs, MkPs, and ErPs are highly enriched for RUNX1 target genes (Figure 1A). Chromatin immunoprecipitation (ChIP) enrichment analysis by Enrichr17 of DEGs revealed that potential RUNX1 target genes are enriched in genes that are lower in MEPs than CMPs (adjusted P = 1.02e−04), more highly expressed in MkPs than MEPs (adjusted P = 6.30e−06), and have lower expression in ErPs than MEPs (adjusted P = 6.22e−13). Similarly, when DEGs from new bulk RNA-seq data sets were clustered based on expression differences between all 4 populations, the clusters that had genes upregulated in MkPs and downregulated in ErPs compared with MEPs (supplemental Figure 1A, groups 4 and 5; supplemental Table 1) were enriched for RUNX1 target genes. RUNX1 targets were not enriched in other clusters.
RUNX1 activity promotes megakaryocytic fate in human MEPs. (A) Summary of human progenitor populations analyzed (left) and data predicting that RUNX1 regulates the differential gene expression between these populations (right). Shown are the number of DEGs that are increased (red) or decreased (blue) in expression between population pairs. Number of predicted RUNX1 target genes and adjusted P values from Enrichr. (B) Effects on colony formation of MEPs transduced with retrovirus encoding full-length RUNX1 or empty vector control. (C) Effects on colony formation of MEPs treated with DMSO (control) or 2 μM Ro5-3335 for 48 hours before plating for colony formation in the absence of drug. (D) Effects on colony formation of MEPs treated with 50 nM all-trans retinoic acid (ATRA) or 50 nM Rapamycin (Rapa), with or without 2 μM Ro5-3335 (Ro5) for 48 hours before plating for colony formation in the absence of drug. Colony types were determined based on CD41 and CD235 immunofluorescence. Mk/E, colonies contain both erythroid and megakaryocytic cells, Mk, megakaryocyte only colonies, E, erythroid only colonies. Each figure represents 3 individual experiments. Average ± standard deviation (SD) are graphed (∗P < .05, ∗∗P < .01). Data from individual experiments provided in supplemental Figure 2.
RUNX1 activity promotes megakaryocytic fate in human MEPs. (A) Summary of human progenitor populations analyzed (left) and data predicting that RUNX1 regulates the differential gene expression between these populations (right). Shown are the number of DEGs that are increased (red) or decreased (blue) in expression between population pairs. Number of predicted RUNX1 target genes and adjusted P values from Enrichr. (B) Effects on colony formation of MEPs transduced with retrovirus encoding full-length RUNX1 or empty vector control. (C) Effects on colony formation of MEPs treated with DMSO (control) or 2 μM Ro5-3335 for 48 hours before plating for colony formation in the absence of drug. (D) Effects on colony formation of MEPs treated with 50 nM all-trans retinoic acid (ATRA) or 50 nM Rapamycin (Rapa), with or without 2 μM Ro5-3335 (Ro5) for 48 hours before plating for colony formation in the absence of drug. Colony types were determined based on CD41 and CD235 immunofluorescence. Mk/E, colonies contain both erythroid and megakaryocytic cells, Mk, megakaryocyte only colonies, E, erythroid only colonies. Each figure represents 3 individual experiments. Average ± standard deviation (SD) are graphed (∗P < .05, ∗∗P < .01). Data from individual experiments provided in supplemental Figure 2.
To investigate whether RUNX1 promotes Mk fate, we overexpressed RUNX1 in MEPs by retroviral transduction. Notably, there are 3 RUNX1 isoforms; RUNX1B and RUNX1C are active transcriptionally, whereas RUNX1A, which lacks the transcriptional activation domain, is inactive. RUNX1C is transcribed from the distal promoter and is 27 amino acids longer than RUNX1B (supplemental Figure 1B). In mice, proximal promoter usage is only observed in committed MkPs whereas the distal promoter is active in adult hematopoietic stem and progenitor cells.18 Because our RNA-seq data show that RUNX1C expression is significantly higher than RUNX1B in CMPs, MEPs, and ErPs and that the RUNX1B isoform is not detected in primary human MkPs (supplemental Figure 1C), we used the RUNX1C isoform (hereafter called RUNX1). Overexpression of RUNX1 vs empty vector in MEPs increased Mk-only colonies (P = 3.01e−02; Figure 1B; supplemental Figure 2A).
To further investigate the role of RUNX1 in MEP fate specification, we used short-hairpin RNA (sh-RNA) to knock down RUNX1 expression but did not observe a significant change in colony outcomes. Because RUNX1 knockdown can induce compensatory increases of RUNX2/3 expression,19 which may account for the absence of phenotype, we inhibited the activity of all RUNX family proteins using Ro5-3335, an indirectly acting small molecule inhibitor widely used in the field. MEPs treated with 2 μM Ro5 for 48 hours had increased E-only colonies (P = 7.09e−03; Figure 1C; supplemental Figure 2B). Use of the more specific but less potent pan-RUNX inhibitor, AI-14-91, which directly inhibits heterodimerization of RUNX family proteins with CBFβ,14 also resulted in increased E-only colonies compared with controls (P = 1.62e−02; supplemental Figure 2C-D), consistent with the hypothesis that RUNX1 promotes Mk fate in MEPs.
We showed previously that drugs and gene knockdowns that decrease cell cycling bias the MEP fate decision5 toward Mk over E commitment. Here, we found that cotreatment with the RUNX1 inhibitor Ro5-3335 reverses the Mk bias induced by rapamycin or all-trans retinoic acid, and promotes E fate in MEPs (Figure 1D; supplemental Figure 2E) suggesting that RUNX1 acts downstream of decreased cell cycling to promote the Mk bias of MEPs.
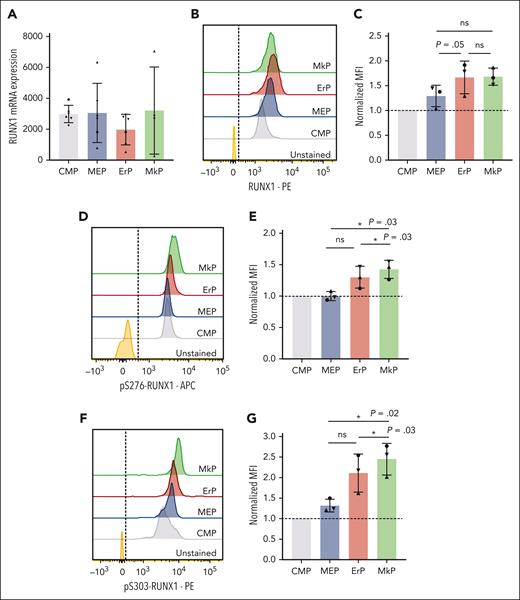
MkPs and ErPs have similar RUNX1 RNA and protein levels
Based on known effects of RUNX1 in Mk maturation20,21 and our data showing that RUNX1 promotes Mk commitment of MEPs, we assessed whether RUNX1 levels increase upon Mk fate commitment. However, RUNX1 messenger RNA levels did not differ significantly between MEPs and upstream (CMPs) and downstream (MkPs and ErPs) progenitor populations (Figure 2A). Using intracellular flow cytometry, mean fluorescence intensity (MFI) of total RUNX1 protein staining showed a trend toward increased RUNX1 protein levels from CMPs to MEPs to downstream progenitors. But the MFI (normalized to CMPs, n = 3 experiments) showed no statistically significant differences in total RUNX1 protein between MkPs and ErPs (Figure 2B-C).
MkPs and ErPs have similar RUNX1 RNA and protein levels, but pS-RUNX1 levels are highest in MkPs. (A) DESeq2-normalized counts between CMPs, MEPs, ErPs, and MkPs were compared from 6 biological replicates (4 for MkPs). Dots of the same shape represent data from the same donor. (B,D,F) Representative images of intracellular staining with antibody targeting total RUNX1 protein (B), pS276-RUNX1 (D), and pS303-RUNX1 (F) with unstained cells used as controls. Dashed line is the cutoff for positive staining. (C,E,G) y-axes: the MFI for each population after normalization to the MFI of CMPs in each experiment. Dots of the same shape represent 1 experiment (from the same donor, processed in parallel). Average ± SD shown (∗P < .05). ns, not significant.
MkPs and ErPs have similar RUNX1 RNA and protein levels, but pS-RUNX1 levels are highest in MkPs. (A) DESeq2-normalized counts between CMPs, MEPs, ErPs, and MkPs were compared from 6 biological replicates (4 for MkPs). Dots of the same shape represent data from the same donor. (B,D,F) Representative images of intracellular staining with antibody targeting total RUNX1 protein (B), pS276-RUNX1 (D), and pS303-RUNX1 (F) with unstained cells used as controls. Dashed line is the cutoff for positive staining. (C,E,G) y-axes: the MFI for each population after normalization to the MFI of CMPs in each experiment. Dots of the same shape represent 1 experiment (from the same donor, processed in parallel). Average ± SD shown (∗P < .05). ns, not significant.
pS-RUNX1 levels are higher in MkPs than other progenitors
Posttranslational modification of RUNX1 affects its function and stability.22 Intracellular flow cytometry using specific antibodies against pS276 and pS303 that are required for the in vitro transcriptional activity of RUNX123 revealed that MkPs have significantly higher pS276- and pS303-RUNX1 levels than CMPs, MEPs, or ErPs (Figure 2D-G).
Phosphomimetic RUNX1-4D promotes Mk fate bias in MEPs and HEL cell line
Because our analyses suggest that Mk commitment is associated with increased pS-RUNX1, we tested whether changes in the RUNX1 phosphorylation state differentially affect MEP fate. We used previously described13 RUNX1 constructs mutated at S276, S293, T300, and S303 (Figure 3A). RUNX1-4A has mutations to alanine, preventing phosphorylation of the 4 sites, and RUNX1-4D has mutations to aspartic acid, mimicking phosphorylated residues. Whereas enforced expression of RUNX1-4A does not, both RUNX1-WT and RUNX1-4D cause a statistically significant increase in Mk-only colonies and RUNX-4D significantly decreases E-only colonies (Figure 3B; supplemental Figure 2F).
Phosphomimetic RUNX1-4D promotes Mk fate bias in MEPs and the HEL cell line. (A) Schematic of HA-tagged RUNX1-WT, RUNX1-4A, and RUNX1-4D constructs. Four S/T sites downstream of the DNA binding domain are mutated to alanine [4A] or aspartic acid [4D]. HA-tag is at the N-terminus. Additional serine phosphorylation sites are also highlighted. (B) Effects on colony formation of MEPs transduced with transgenes indicated. Each figure represents 3 individual experiments. Colony types as described in Figure 1. Data from individual experiments provided in supplemental Figure 2G. (C) Western blot probed with anti-HA antibody to detect transgenic proteins in transduced HEL cells. (D) Bar graph representing the mean and SD (error bars) of anti-HA western blot data from 3 biological repeats. Band intensities were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) signal in the corresponding lanes, then normalized to RUNX1-WT (dashed line, 1.0). Dots of the same shape represent data from individual experiments. (E) Representative image of CD41/CD42 staining in HEL cells expressing empty vector (EV) or RUNX1-4A/WT/4D without TPA treatment. (F) Graph with CD41+CD42− population on the bottom and CD41+CD42+ on top (n = 5). For panels B,D,F, average ± SD are graphed (∗P < .05, ∗∗∗P < .001, ∗∗∗∗P < .0001). Data from individual experiments provided in supplemental Figure 3D.
Phosphomimetic RUNX1-4D promotes Mk fate bias in MEPs and the HEL cell line. (A) Schematic of HA-tagged RUNX1-WT, RUNX1-4A, and RUNX1-4D constructs. Four S/T sites downstream of the DNA binding domain are mutated to alanine [4A] or aspartic acid [4D]. HA-tag is at the N-terminus. Additional serine phosphorylation sites are also highlighted. (B) Effects on colony formation of MEPs transduced with transgenes indicated. Each figure represents 3 individual experiments. Colony types as described in Figure 1. Data from individual experiments provided in supplemental Figure 2G. (C) Western blot probed with anti-HA antibody to detect transgenic proteins in transduced HEL cells. (D) Bar graph representing the mean and SD (error bars) of anti-HA western blot data from 3 biological repeats. Band intensities were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) signal in the corresponding lanes, then normalized to RUNX1-WT (dashed line, 1.0). Dots of the same shape represent data from individual experiments. (E) Representative image of CD41/CD42 staining in HEL cells expressing empty vector (EV) or RUNX1-4A/WT/4D without TPA treatment. (F) Graph with CD41+CD42− population on the bottom and CD41+CD42+ on top (n = 5). For panels B,D,F, average ± SD are graphed (∗P < .05, ∗∗∗P < .001, ∗∗∗∗P < .0001). Data from individual experiments provided in supplemental Figure 3D.
To dissect the differential effects of RUNX1-4A, RUNX1-4D, and RUNX1-WT at the epigenetic level, we expressed HA-tagged RUNX1-4A, RUNX1-WT, and RUNX1-4D in HEL cells, which have some functional and immunophenotypic features of MEPs.24-27 Anti-HA western blots (Figure 3C-D) showed that the RUNX1-WT transgene was expressed at a higher level than RUNX1-4A and RUNX1-4D. Western blot analysis using anti-RUNX1 to determine the degree to which enforced expression of the RUNX1 constructs changed total RUNX1 protein levels (endogenous plus exogenous) revealed that total RUNX1 protein in RUNX1-WT HEL cells was ∼2.5-fold higher than empty vector controls. Enforced RUNX1-4A and RUNX1-4D transgene expression led to RUNX levels 1.3-fold higher than controls (supplemental Figure 3A-B). Because there have been reports of RUNX1 mutants that have altered subcellular localization,28 immunofluorescence was performed, which confirmed the nuclear localization of all 3 RUNX1 transgenes in HEL cells (supplemental Figure 3C).
Enforced RUNX1-WT and RUNX1-4D expression affected the surface antigen phenotype of HEL cells. HEL cells express the megakaryocytic surface marker CD41 in control culture conditions, but do not express CD42, a marker of Mk maturation, unless differentiation is induced with phorbol ester (TPA).24,26,27 Even in the absence of TPA, there was a significant increase in surface CD42 protein with enforced expression of RUNX1-WT and RUNX1-4D but not with RUNX1-4A (Figure 3E-F). RUNX1-4D promoted significantly more CD42 than RUNX1-WT (supplemental Figure 3D). RUNX1-4D–expressing HEL cells also expressed CD49f, a different Mk maturation marker, at higher levels than those expressing RUNX1-4A or RUNX1-WT without TPA treatment (supplemental Figure 3E-F). In response to TPA, HEL cells expressing RUNX1-4A, RUNX1-WT, or RUNX1-4D showed equivalent expression of CD42 and CD49f, indicating that RUNX1-4A does not block TPA-induced HEL cell maturation (supplemental Figure 3G-J).
RUNX1 chromatin association is minimally affected by RUNX1-4A and RUNX1-4D
Anti-HA CUT&RUN to assess differential chromatin localization of the RUNX1 variants in HEL cells revealed that RUNX1-4A, RUNX1-WT, and RUNX1-4D had 5815, 6567, and 5110 peaks, respectively, that were conserved between 2 biological replicates (supplemental Figure 4A). Motif analysis by STREME29 revealed high enrichment of the RUNX1 consensus binding domain (TGTGGTT) for each of the constructs (Figure 4A). The peaks were also enriched for binding motifs of ETS and GATA that are known to interact with RUNX1.30,31 Many (41%-46%) of the conserved peaks were within 1 kilobase (kb) of the transcription start site (TSS) of the nearest gene (Figure 4B). When these conserved peaks were compared with publicly available data sets in Cistrome-DB,32 the peaks were colocalized with various transcription factors (supplemental Figure 4B) with CBFβ, the essential RUNX1 cofactor, at the top of the list. Our data set correlates significantly with a previous RUNX1 ChIP-sequencing (ChIP-seq) data set performed on primary human megakaryocytes derived from cord blood (supplemental Figure 4B, red dots),33 validating HEL cells as a model for assessing RUNX1 chromatin association in human hematopoietic cells. In addition, transcription factors known to bind to the same regions as RUNX1 as part of combinatorial transcriptional control (eg, GATA2, ERG, and FLI1)34 show high correlation scores. Other transcription factors that overlap significantly with RUNX1 in their DNA binding sites, for example LEF1, could also be cooperating with RUNX1 to regulate fate specification.
RUNX1 chromatin association is not significantly affected by RUNX1-4A and RUNX1-4D. (A) STREME (simple, thorough, rapid, enriched motif elicitation) motif enrichment analysis showing that CUT&RUN binding sites for RUNX1-4A, RUNX1-WT, and RUNX1-4D are highly enriched for the conserved RUNX1 binding motif, as well as ETS and GATA binding motifs. (B) Distance to the nearest TSS for RUNX1-4A (4A), RUNX1-WT (WT), and RUNX1-4D (4D) binding sites. (C) Differential binding analysis of RUNX1 mutants compared with WT (4A: left, 4D: middle). Cutoff for significance was adjusted P < .05 (horizontal dashed line), and log2 fold change (log2FC) of less than −1 or >1 (vertical dashed lines). Log2 fold change of <0 indicates that RUNX1-WT peak is higher than that of RUNX1-4A or RUNX1-4D. Differential binding analysis of RUNX1-4A vs RUNX1-4D (right) shows 127 peaks beyond the significance cutoff in red.
RUNX1 chromatin association is not significantly affected by RUNX1-4A and RUNX1-4D. (A) STREME (simple, thorough, rapid, enriched motif elicitation) motif enrichment analysis showing that CUT&RUN binding sites for RUNX1-4A, RUNX1-WT, and RUNX1-4D are highly enriched for the conserved RUNX1 binding motif, as well as ETS and GATA binding motifs. (B) Distance to the nearest TSS for RUNX1-4A (4A), RUNX1-WT (WT), and RUNX1-4D (4D) binding sites. (C) Differential binding analysis of RUNX1 mutants compared with WT (4A: left, 4D: middle). Cutoff for significance was adjusted P < .05 (horizontal dashed line), and log2 fold change (log2FC) of less than −1 or >1 (vertical dashed lines). Log2 fold change of <0 indicates that RUNX1-WT peak is higher than that of RUNX1-4A or RUNX1-4D. Differential binding analysis of RUNX1-4A vs RUNX1-4D (right) shows 127 peaks beyond the significance cutoff in red.
Differential binding analysis of the peaks for each RUNX1 transgene revealed extensive overlap between chromatin association of RUNX1-WT, RUNX1-4A, and RUNX1-4D. Although neither RUNX1-4A nor RUNX1-4D showed significant differences in binding compared with RUNX1-WT, RUNX1-4A vs RUNX1-4D showed differential binding at 127 peaks (Figure 4C); 45 peaks were more significantly bound by RUNX1-4A, whereas 82 peaks (at 60 genes) were more significantly bound by RUNX1-4D. We assessed whether the genes proximal to these differentially bound peaks are differentially expressed in RUNX1-4A– vs RUNX-4D–expressing cells, or in primary human MEPs vs MkPs; 25 genes had differential expression in RUNX1-4A– vs RUNX1-4D–expressing cells. Genes proximal to peaks with higher RUNX1-4D binding tended to show higher expression in RUNX1-4D–expressing cells (supplemental Table 4). Assessment of gene expression in primary cells revealed that 6 genes were differentially expressed in MEPs vs MkPs, with no correlation in binding intensity and expression levels.
HEL cells expressing RUNX1-4A/WT/4D reveal distinct gene expression patterns
RNA-seq analysis of HEL cells expressing WT and mutant RUNX1 identified 2625 DEGs with an adjusted P value of P < .05 (Figure 5A). The replicates for each sample clustered closely by principal component analysis after batch effect correction (supplemental Figure 5A). Using DEGreport,35 the 2 largest gene expression clusters showed strong concordance for the effects of RUNX1-WT and RUNX1-4D with either decreased (cluster 1) or increased (cluster 2) expression compared with empty vector and RUNX1-4A–expressing cells (Figure 5B, gene cluster lists; supplemental Table 2). This is consistent with data in primary MEPs and HEL cells in which RUNX1-WT and RUNX1-4D induce a more robust phenotype than RUNX1-4A.
HEL cells expressing RUNX1-WT/4A/4D reveal distinct gene expression patterns. (A) Heat map of 2625 most DEGs (adjusted P < .05) between the 3 transgene expressing HEL cells. (B) Gene expression patterns of DEGs. Most (∼80%) DEGs were grouped into the 2 largest clusters showing RUNX1-WT and RUNX1-4D decreasing (cluster 1, 1066 genes) or increasing (cluster 2, 1003 genes) expression over EV and RUNX1-4A. (C) Location of RUNX1 binding relative to the TSS for the 1550 DEGs with an associated RUNX1 CUT&RUN peak shared by all 3 (RUNX1-4A/WT/4D) transgenes. (D-E) Representative genes with increased (D, GP1BB) or decreased (E, KIT) expression by RUNX1-WT and RUNX1-4D compared with EV and RUNX1-4A with peaks near TSS. Top rows show 2 replicates of CUT&RUN for each transgene, and bottom rows show 2 replicates of corresponding RNA-seq data. (F) Plots showing DEGs between MkPs vs MEPs and ErPs vs MEPs from bulk RNA-seq data on y-axis and x-axis, respectively, with genes from cluster 1 (left, in red) and cluster 2 (right, in green) highlighted. For all DEGs, the P value cutoff was .05, and log2FC cutoff was 1. Genes that met the P value cutoff but not log2FC cutoff are localized to gray box. Number of genes that overlap with clusters 1 or 2 out of total number of DEGs, along with P values calculated by hypergeometric test are shown in each quadrant.
HEL cells expressing RUNX1-WT/4A/4D reveal distinct gene expression patterns. (A) Heat map of 2625 most DEGs (adjusted P < .05) between the 3 transgene expressing HEL cells. (B) Gene expression patterns of DEGs. Most (∼80%) DEGs were grouped into the 2 largest clusters showing RUNX1-WT and RUNX1-4D decreasing (cluster 1, 1066 genes) or increasing (cluster 2, 1003 genes) expression over EV and RUNX1-4A. (C) Location of RUNX1 binding relative to the TSS for the 1550 DEGs with an associated RUNX1 CUT&RUN peak shared by all 3 (RUNX1-4A/WT/4D) transgenes. (D-E) Representative genes with increased (D, GP1BB) or decreased (E, KIT) expression by RUNX1-WT and RUNX1-4D compared with EV and RUNX1-4A with peaks near TSS. Top rows show 2 replicates of CUT&RUN for each transgene, and bottom rows show 2 replicates of corresponding RNA-seq data. (F) Plots showing DEGs between MkPs vs MEPs and ErPs vs MEPs from bulk RNA-seq data on y-axis and x-axis, respectively, with genes from cluster 1 (left, in red) and cluster 2 (right, in green) highlighted. For all DEGs, the P value cutoff was .05, and log2FC cutoff was 1. Genes that met the P value cutoff but not log2FC cutoff are localized to gray box. Number of genes that overlap with clusters 1 or 2 out of total number of DEGs, along with P values calculated by hypergeometric test are shown in each quadrant.
Integration of RNA-seq and CUT&RUN data
Of 2625 DEGs in RUNX1-4A/WT/4D–expressing HEL cells, 1550 (59.0%) were the nearest gene to at least 1 of the peaks shared by RUNX1-4A/WT/4D, with 953 genes having a shared peak within 1 kb of the TSS (Figure 5C; supplemental Table 2). Cluster 2 genes, showing higher expression in RUNX1-WT/4D–expressing HEL cells, that have RUNX1 peaks in their promoters are enriched for genes associated with megakaryopoiesis and platelet activation including GP6, GP9, GP1BA, GP1BB, ITGA2, and ITGA6 (Figure 5D; supplemental Figure 5B-C).36-38 Genes that are required for erythropoiesis (KIT and ID2) and those that need to be downregulated for megakaryopoiesis (ANKRD26) are downregulated more by RUNX1-WT and RUNX1-4D than RUNX1-4A (Figure 5E; supplemental Figure 5D-E).39,40 Data mining for other genes that are lower in ErPs than MEPs (supplemental Table 1) and decreased by RUNX1-WT and RUNX1-4D in HEL cells with a proximal RUNX1 peak (eg, SOCS2) may identify novel regulators of erythroid maturation.
We compared the DEGs between RUNX1-4A–, RUNX1-WT–, and RUNX1-4D–expressing HEL cells with the DEGs between primary human MEPs, MkPs, and ErPs (Figure 5F). The 1523 genes upregulated in MkPs vs MEPs (top quadrants), were significantly enriched for genes in HEL cluster 2 (P = 4.67e−22) but not HEL cluster 1 (P = .32, P value calculated by hypergeometric test with genes that have average DESeq2-normalized counts of >8, rationale for cutoff provided in supplemental Figure 5F).41 Of 1467 genes downregulated in MkPs vs MEPs (bottom quadrants), genes from cluster 1 were significantly enriched (P = 2.93e−25) but not those from cluster 2 (P = .11). Thus, the genes with increased expression upon RUNX1 phosphorylation in HEL cells overlap with genes upregulated during megakaryocytic fate specification of MEPs, whereas the downregulated genes overlap with those downregulated during MEP to MkP differentiation. The association with genes differentially expressed between ErPs and MEPs was less significant than between MkP and MEP. These results support the hypothesis that phosphorylated-serine/-threonine RUNX1 drives MEP fate toward the Mk lineage by acting as both a transcriptional activator and repressor for distinct target genes.
CDK9 phosphorylates RUNX1, and CDK9 degradation increases erythroid commitment in MEPs
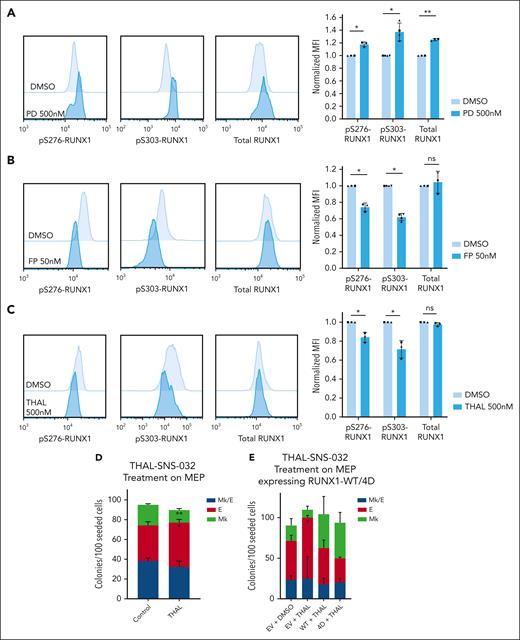
Multiple kinases have been reported to phosphorylate RUNX1 S/T residues including extracellular signal-regulated kinase, homeodomain-interacting protein kinase 2 (HIPK2), and CDKs 1, 2, and 6.13,42-44 We tested whether inhibition of these kinases affects pS-RUNX1 in MEPs. Surprisingly, neither sh-RNAs targeting HIPK2 or MAPK1 (supplemental Figure 6A) nor commercially available CDK1/2 inhibitors (BMS265246, 50% inhibitory concentration = 6 nM and 9 nM for CDK1 and 2, respectively) significantly affected pS-RUNX1 levels (supplemental Figure 6B-C) except that HIPK2 knockdown caused a consistent but small decrease in pS276-RUNX1. The CDK4/6 inhibitor palbociclib (PD0332991, 50% inhibitory concentration = 11 nM and 16 nM for CDK4 and 6, respectively) actually increased pS-RUNX1 and total RUNX1 levels in MEPs (Figure 6A) consistent with its promotion of Mk fate. Thus, none of the previously reported kinases appears to affect RUNX1 phosphorylation in MEPs.
CDK9 degradation reduces S276/S303-RUNX1 phosphorylation and promotes erythroid fate specification in MEPs. (A-C) Representative images of intracellular staining of pS276-RUNX1, pS303-RUNX1, and total RUNX1 (left) in DMSO vs 500 nM Palbociclib (PD 0332991, PD) for 48 hours (A), DMSO vs 50 nM flavopiridol (FP) for 2 hours (B), and DMSO vs 500 nM THAL-SNS-032 (THAL) for 16 hours (C). Bars show average ± SD MFI after normalization to the MFI of DMSO in each experiment (right). Dots of the same shape represent 1 experiment (from the same donor, processed in parallel). (D) Effects on colony formation of MEPs treated with DMSO or 500 nM THAL-SNS-032 for 48 hours. Average ± SD shown (n = 3). (E) Colony formation of MEPs transduced to express RUNX1-WT or RUNX1-4D, with or without 500 nM THAL treatment for 48 hours. For all plots, ∗P < .05, ∗∗P < .01.
CDK9 degradation reduces S276/S303-RUNX1 phosphorylation and promotes erythroid fate specification in MEPs. (A-C) Representative images of intracellular staining of pS276-RUNX1, pS303-RUNX1, and total RUNX1 (left) in DMSO vs 500 nM Palbociclib (PD 0332991, PD) for 48 hours (A), DMSO vs 50 nM flavopiridol (FP) for 2 hours (B), and DMSO vs 500 nM THAL-SNS-032 (THAL) for 16 hours (C). Bars show average ± SD MFI after normalization to the MFI of DMSO in each experiment (right). Dots of the same shape represent 1 experiment (from the same donor, processed in parallel). (D) Effects on colony formation of MEPs treated with DMSO or 500 nM THAL-SNS-032 for 48 hours. Average ± SD shown (n = 3). (E) Colony formation of MEPs transduced to express RUNX1-WT or RUNX1-4D, with or without 500 nM THAL treatment for 48 hours. For all plots, ∗P < .05, ∗∗P < .01.
To identify the kinase that phosphorylates RUNX1 in MEPs, we used a kinase prediction tool to obtain lists of kinases likely to phosphorylate the 4 S/T sites mutated in RUNX1-4A and RUNX1-4D (supplemental Table 3).45 Although various CDKs were highly ranked in the lists, we focused on CDK9 because CDK9 inhibition decreases megakaryocyte maturation, induces erythroid differentiation, and reduces activity of the RUNX1-CBFβ-GATA1 complex in K562 cell lines.46,47 Although partial CDK9 knockdown by sh-RNA did not affect RUNX1 phosphorylation level (supplemental Figure 6D-E), flavopiridol treatment significantly decreases pS276-RUNX1 and pS303-RUNX1 without affecting total RUNX1 levels (Figure 6B). Flavopiridol, a pan-CDK inhibitor, inhibits CDK1, 2, 4, 6, 7, and 9. Our assays showed that CDK1, 2 (BMS265246), 4 and 6 (palbociclib) inhibition does not decrease RUNX1 phosphorylation and the flavopiridol concentration we used does not affect CDK7 activity.48 To further test the effect of CDK9 decrease on RUNX1 phosphorylation, we used THAL-SNS-032, a selective CDK9 degrader (Protac) to induce significant CDK9 decrease in MEPs with 500 nM selected because it decreases CDK9 and is not toxic to MEPs with up to 48-hour treatment (supplemental Figure 7A). THAL-SNS-032 treatment of MEPs decreased pS276- and pS303-RUNX1 but not total RUNX1 levels, suggesting that CDK9 phosphorylates RUNX1 in primary MEPs (Figure 6C). We tested the effect of CDK9 downregulation on MEP specification. Treating MEPs with THAL-SNS-032 before plating for CFU assays decreased Mk and promoted E fate (Figure 6D; supplemental Figure 7B) consistent with CDK9-mediated phosphorylation of RUNX1 in MEPs promoting Mk fate. The erythroid fate bias induced by THAL-SNS-032 was prevented by overexpression of RUNX1-4D, but not RUNX1-WT, strongly suggesting that CDK9-mediated phosphorylation of RUNX1 is critical for this effect. (Figure 6E; supplemental Figure 7C-D).
Discussion
RUNX1 has been widely studied for its role in early hematopoietic development, megakaryocyte maturation, and AML. However, the role of RUNX1 in megakaryocytic vs erythroid fate specification at the level of primary bipotent progenitors has not been explored previously. We show in primary human MEPs that phosphorylated-serine/-threonine RUNX1 promotes megakaryocytic at the expense of erythroid fate. We also show that RUNX1 differentially activates Mk-lineage genes and inhibits E-lineage genes in the HEL cell line depending on its phosphorylation status, and that the DEGs in HEL cells overlap significantly with those that are differentially expressed between primary MEPs and MkPs, which provides insight into the gene regulatory mechanisms underlying the phenotype observed in MEPs.
Although endogenous RUNX1 protein and RNA levels are not increased in MkPs vs ErPs, pS-RUNX1 levels are significantly higher in MkPs than ErPs. To confirm that phosphorylation of RUNX1 is critical for its function in promoting Mk fate, we show that RUNX1-4D, a phosphomimetic mutant, induces megakaryocytic gene expression and phenotypic changes that are diminished or absent with RUNX1-4A. The trend toward enhanced Mk fate by RUNX1-4A may be because of residual activity from remaining unmodified serines including S48, S424, and S462. Although the 4 S/T residues that were mutated in our 4A/4D mutants (S276, S293, T300, and S303) have the strongest effects on RUNX1 activity,23 phosphorylation of additional S/T residues also contributes to RUNX1 activity. For example, to block hematopoiesis in mice, phosphorylation of RUNX1 S462 needs to be prevented in addition to the 4 S/T residues mutated in RUNX1-4A.49 Also, phosphomimetic mutations of S48, S303, and S424 enhance RUNX1 transcriptional activity.15
Kuvardina et al11 reported that RUNX1 represses KLF1 during megakaryocytic differentiation of human CD34+ cells, and RUNX1 binding to KLF1 promoter in K562 cells increases with TPA treatment. KLF1 activates multiple erythroid genes and has a crossantagonistic relationship with FLI1.50 However, KLF1 and FLI1 were not differentially expressed in HEL cells with induced expression of RUNX1-4A/WT/4D. The difference in cell line models and TPA-induced differentiation likely account for these disparities. Notably, KLF1 is expressed in HEL cells and there is a region ∼4 kb upstream of KLF1 that binds all 3 forms of RUNX1.
Our CUT&RUN data from HEL cells show that chromatin association of RUNX1 occurs predominantly near TSS. Enrichment analyses of the RUNX1-4A/WT/4D peaks show significant similarity to the published anti-RUNX1 ChIP-seq data conducted using in vitro differentiated human megakaryocytes.33 In published anti-RUNX1 ChIP-seq data from TPA-induced K562 cells, >50% RUNX1 peaks were found >5 kb from TSS, but there was also a high peak concentration near TSS.51 Differences are likely because of the use of TPA or to differences between cell lines. Despite partial discrepancy, RUNX1-associated regions in TPA-treated K562 cells51 are enriched in RUNX1, GATA, and ETS motifs.
It is important to consider the context of transgene overexpression in the HEL cells because endogenous RUNX1 is expressed. The DEGs that have increased or decreased expression compared with control HEL cells indicate that RUNX1-WT and RUNX-4D have additive effects over endogenous RUNX1. For a more explicit comparison of RUNX1-4A/WT/4D function, RUNX1-knockout cell lines could be generated. The difference in transgene expression levels is another limitation for these studies. RUNX1-4A and RUNX1-4D are expressed at a lower level than RUNX1-WT. When expressed at similar levels, some transgene-induced DEGs might show more dramatic expression changes. A previous report suggests that RUNX1-4A is more stable and RUNX1-4D less stable than RUNX1-WT.13 This pattern, not observed in our HEL cell lines, could be cell-type specific.
Our novel finding that CDK9 can phosphorylate RUNX1 adds context to the previous findings, suggesting a direct link between CDK9- and RUNX1-mediated transcriptional changes. Together with cyclin T1, CDK9 forms the P-TEFb complex, which phosphorylates serine 2 of the RNA polymerase 2 C-terminal domain to facilitate the transition from transcriptional initiation to elongation.52 Jiang et al showed that RUNX1 and cyclin T1, expressed in 293T cells with tags, are coimmunoprecipitated, confirming the interaction of RUNX1 and P-TEFb complexes.53 In prior studies,46 CDK9 inhibition with flavopiridol decreased Mk output from CD34+ cells. In K562 cells,47 the CDK9 inhibitor Wogonin caused a dose-dependent increase of erythroid markers. Elagib et al46 showed that P-TEFb inhibition decreases the transcriptional synergy between GATA1, RUNX1, and CBFβ, but the mechanisms underlying this association of CDK9 with megakaryopoiesis were not clear. Possibly, when CDK9 phosphorylates RUNX1, it alters RUNX1’s affinity to transcription cofactors, leading to enhanced activity.
Investigating the binding partners that bind differentially to RUNX1-4A/WT/4D could provide more insight into RUNX1’s differential activity in response to S/T phosphorylation. Guo et al showed that RUNX1 phosphorylated at S48, S303, and S424 has decreased histone deacetylase association and is thus potentially less inhibitory.54 If this also holds true for our RUNX1-4D transgene, less histone deacetylase association, and the subsequent changes in chromatin that lead to increased transcription could in part underlie the increased expression of megakaryocytic genes.
In conclusion, we have discovered that phosphorylated-serine/-threonine RUNX1 promotes megakaryocytic fate in primary human MEPs and induces megakaryocytic features in the HEL cell line. Although mutations in 4 central RUNX1 residues that mimic or block phosphorylation do not significantly affect DNA binding, the phosphomimetic construct more potently induces megakaryocytic fate. We have also discovered that CDK9 can phosphorylate RUNX1 and that inhibiting CDK9 affects MEP fate specification.
Acknowledgments
The authors thank Alan Friedman for providing pS303-RUNX1 antibody and for helpful discussions. The authors also thank Dong-Er Zhang for providing the retroviral expression vectors. High throughput sequencing was performed by Genomics and Bioinformatics Core at Yale Stem Cell Center and Yale Center for Genomic Analysis. The authors are grateful to the staff in Yale Flow Cytometry Facility for their support with cell sorting.
This work was funded, in part, by National Institutes of Health (NIH), National Institute of Diabetes and Digestive and Kidney Diseases grants R01 DK114031, R01 DK094934, and RC2 DK122376, as well as U54 DK106857 for the Yale Cooperative Center of Excellence in Hematology. This work was also supported by Chen Innovation Fund for Stem Cell Research (GS100551). Storage and analysis of the sequence data was supported by HPC (High Performance Computing) equipment grant (1S10OD030363-01A1) from the NIH, Office of the Director. The Yale Flow Cytometry Facility is funded by NIH, National Cancer Institute grant P30 CA016359. N.K. received support from the American Heart Association Predoctoral Fellowship (915753). E.N.T. is supported by NIH, National Heart, Lung, and Blood Institute grant F31 HL167628.
N.K. and E.N.T. are PhD candidates at Yale University. This work is submitted in partial fulfillment of the requirement for the PhD.
Authorship
Contribution: N.K. designed and conducted experiments, analyzed data, and wrote the manuscript; Y.-C.L. designed and conducted experiments and analyzed data; E.N.T. conducted experiments and analyzed data; R.I.M., L.W., and P.-X.Z. sorted primary cells and performed experiments; and D.S.K. designed the study, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Diane S. Krause, Department of Laboratory Medicine, Yale Stem Cell Center, Yale University School of Medicine, 10 Amistad St, New Haven, CT 06519; email: diane.krause@yale.edu.
References
Author notes
N.K. and Y.-C.L. contributed equally to this study.
Presented, in part, in poster form at the International Society for Experimental Hematology, Edinburgh, United Kingdom, 3 September 2022; and the International Society for Experimental Hematology, Brooklyn, NY, 19 August 2023.
High-throughput data are available at Gene Expression Omnibus under accession number GSE232880.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.



![Phosphomimetic RUNX1-4D promotes Mk fate bias in MEPs and the HEL cell line. (A) Schematic of HA-tagged RUNX1-WT, RUNX1-4A, and RUNX1-4D constructs. Four S/T sites downstream of the DNA binding domain are mutated to alanine [4A] or aspartic acid [4D]. HA-tag is at the N-terminus. Additional serine phosphorylation sites are also highlighted. (B) Effects on colony formation of MEPs transduced with transgenes indicated. Each figure represents 3 individual experiments. Colony types as described in Figure 1. Data from individual experiments provided in supplemental Figure 2G. (C) Western blot probed with anti-HA antibody to detect transgenic proteins in transduced HEL cells. (D) Bar graph representing the mean and SD (error bars) of anti-HA western blot data from 3 biological repeats. Band intensities were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) signal in the corresponding lanes, then normalized to RUNX1-WT (dashed line, 1.0). Dots of the same shape represent data from individual experiments. (E) Representative image of CD41/CD42 staining in HEL cells expressing empty vector (EV) or RUNX1-4A/WT/4D without TPA treatment. (F) Graph with CD41+CD42− population on the bottom and CD41+CD42+ on top (n = 5). For panels B,D,F, average ± SD are graphed (∗P < .05, ∗∗∗P < .001, ∗∗∗∗P < .0001). Data from individual experiments provided in supplemental Figure 3D.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/144/17/10.1182_blood.2024023963/1/m_blood_bld-2024-023963-gr3.jpeg?Expires=1769085780&Signature=ryhJGDyuEF1UIUSTiupH~9TQJc9VMupYp7uTE8rYt93Ec7IniICkO7xwGSggddLBM-WP3KAPNhNxTNwzkmWPoRLly91aknafMEIqknqoSx2tzrIxJw1r7UWTLlYALVekU7kfvPgxGCrUR4MkijeSMwjpaL4EAvQTs-fVQcHz1b2az8-88vQ~Nn~pJyByAVdu7FiyBSZHS8~mkqGrNhcvQUEMQEwuxUNR5uk~Y-irCekGlSGDYTxrFCFPe9mpjyYSeFk1g42nku-seJOuS3zhSfkDGxa8cZu6y5tNBfZ~xxm88KUX6jSz-45kU4j6dM2TsHVFfw5eYRrh8nMSzXh4jw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal