In this issue of Blood, Collinge and colleagues1 explore the biological basis of unbalanced MYC fluorescence in situ hybridization (FISH) loss patterns (see figure, bottom) in diffuse large/high-grade B-cell lymphomas with MYC and BCL2 rearrangements (HGBCL-DH-BCL2). They convincingly demonstrate that these rearrangements typically preserve MYC and result in its upregulation, similar to balanced rearrangements. These findings support the recommendation that unbalanced MYC break-apart FISH patterns be regarded as positive for MYC rearrangement when diagnosing HGBCL-DH-BCL2.
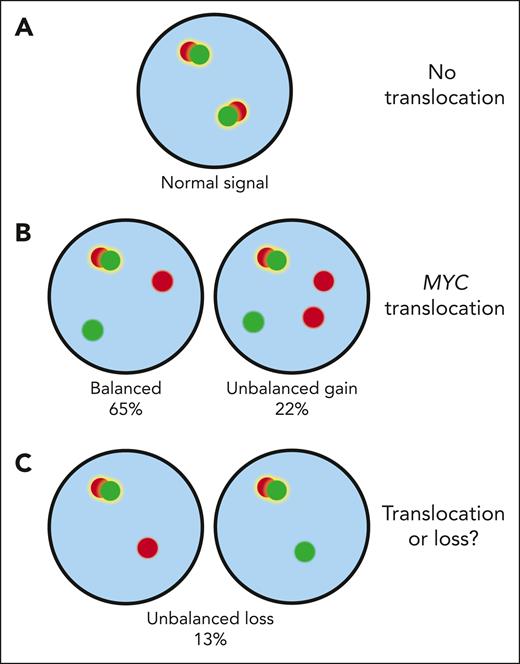
MYC FISH patterns. Cells without a translocation exhibit 2 fusion signals indicative of 2 intact MYC loci (A). In MYC translocated cells, one of the fusion signals is typically split into separate red (centromeric) and green (telomeric) signals. The break-apart can also be accompanied by a gain of the red or green probe (B). So far, there has been uncertainty about the interpretation of unbalanced FISH patterns with loss of either the green or red probe (C). The current study by Collinge et al provides evidence that such alterations reflect genuine rearrangements rather than deletions of the MYC locus.
MYC FISH patterns. Cells without a translocation exhibit 2 fusion signals indicative of 2 intact MYC loci (A). In MYC translocated cells, one of the fusion signals is typically split into separate red (centromeric) and green (telomeric) signals. The break-apart can also be accompanied by a gain of the red or green probe (B). So far, there has been uncertainty about the interpretation of unbalanced FISH patterns with loss of either the green or red probe (C). The current study by Collinge et al provides evidence that such alterations reflect genuine rearrangements rather than deletions of the MYC locus.
HGBCL-DH-BCL2 are aggressive non-Hodgkin lymphomas, which are recognized as a distinct entity in both the current World Health Organization (WHO) Classification of Hematolymphoid Tumors and the International Consensus Classification (ICC). Due to the adverse prognosis associated with HGBCL-DH-BCL2, intensified treatment regimens, such as dose-adjusted etoposide, prednisone, vincristine, cyclophosphamide, doxorubicin, and rituximab (DA-EPOCH-R), are used in many parts of the world.2
Molecular studies support the clinical distinction of HGBCL-DH-BCL2. Almost all cases belong to the germinal center B-cell (GCB) DLBCL subtype and are enriched within the C3/EZB genetic classes.3-6 Recent studies further demonstrate that most HGBCL-DH-BCL2 exhibit a centroblast-like gene expression signature reflective of the dark zone of the germinal center, similar to Burkitt lymphomas.6-8 Interestingly, these signatures are not confined to HGBCL-DH-BCL2, but they identify nearly twice as many aggressive GCB cases lacking double-hit status by FISH, with a dismal prognosis. This suggests that BCL2 and MYC translocations do not fully explain the distinct underlying biology of these lymphomas.
Clinically, identifying HGBCL-DH-BCL2 relies on FISH using break-apart probes to detect the disease-defining translocations. Occasionally, unbalanced MYC break-apart patterns, where the red or green probe signal is lost, are observed. Such patterns could either be explained by rearrangements or by deletions of the MYC locus, leading to uncertainty in classifying these cases (see figure).
This unmet need is addressed in the study by Collinge et al. The authors examined 297 tumors from patients with HGBCL-DH-BCL2 and found that 13% exhibit unbalanced MYC break-apart loss patterns. Among the cases with an unbalanced loss pattern, 85% lacked the telomeric signal (green probe). One hundred thirty cases were evaluable for translocation characterization by sequencing. Among the 17 cases with unbalanced loss patterns, a MYC rearrangement partner could be identified in 12 (71%). The majority involved either immunoglobulin (Ig) loci or other recurrent MYC rearrangement partners. Notably, the MYC gene was consistently preserved on the derivative chromosome, and the authors demonstrated resultant upregulation of MYC at both protein and mRNA levels. These data confirm that such alterations reflect genuine MYC rearrangements rather than deletions. Finally, the authors provide evidence that, like cases with balanced FISH signals, most tumors with loss patterns exhibit the dark zone gene expression signature (68%).
However, some open questions remain. Translocation partners were identified in a smaller fraction of cases with unbalanced rearrangements, and there appear to be more non-Ig translocations than in cases with balanced FISH signals. This indicates heterogeneity in MYC translocations not captured by current methods.9 There is evidence to suggest that the adverse prognosis of HGBCL-DH-BCL2 is confined to cases harboring MYC::Ig translocations.10 Unfortunately, outcome analyses to explore prognostic implications of unbalanced rearrangements remain elusive due to limited case numbers and nonuniform treatment strategies.
The findings presented by Collinge et al have meaningful clinical implications. By demonstrating that unbalanced MYC FISH patterns typically represent bona fide MYC rearrangements, the authors provide a solid rationale for interpreting such signals as positive for MYC translocations. This may enhance diagnostic precision and ensure that patients receive appropriate therapeutic interventions. The study by Collinge and colleagues therefore potentially affects the disease classification of a significant minority of HGBCL-DH-BCL2. Future studies will need to resolve the implications of different types of MYC rearrangements, particularly with regard to its translocation partner, and determine whether gene expression-based classifiers should complement FISH studies in routine clinical workup.
Conflict-of-interest disclosure: S.K.A. reports consultancy for Foresight Diagnostics.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal