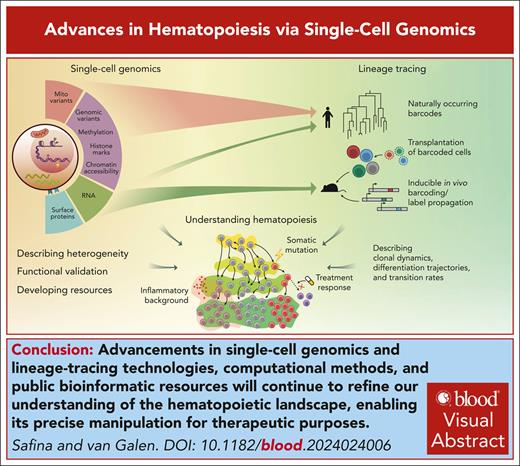
Visual Abstract
Recent advancements in single-cell genomics have enriched our understanding of hematopoiesis, providing intricate details about hematopoietic stem cell biology, differentiation, and lineage commitment. Technological advancements have highlighted extensive heterogeneity of cell populations and continuity of differentiation routes. Nevertheless, intermediate “attractor” states signify structure in stem and progenitor populations that link state transition dynamics to fate potential. We discuss how innovative model systems quantify lineage bias and how stress accelerates differentiation, thereby reducing fate plasticity compared with native hematopoiesis. We conclude by offering our perspective on the current model of hematopoiesis and discuss how a more precise understanding can translate to strategies that extend healthy hematopoiesis and prevent disease.
Introduction
The human body produces >100 billion new blood cells daily through a tightly regulated process called hematopoiesis.1-3 Hematopoiesis has been historically represented as a branched hierarchical process, in which a multipotent hematopoietic stem cell (HSC) at the top of the hierarchy produces all mature cells of the erythroid, megakaryocytic, myeloid, and lymphoid lineages through a set of oligopotent progenitors. This finely tuned process of differentiation replenishes an array of blood cell types ranging from erythrocytes that transport oxygen and megakaryocytes that produce platelets to monocytes and T cells that mediate innate and adaptive immunity. Although the concept of a multipotent stem cell was suggested >100 years ago,4 the experimental development of this model started in the 1960s, when the existence of HSCs was inferred from the observation that transplantation of mouse bone marrow cells gave rise to multilineage clonal colonies in the spleens of recipient mice.5,6 Later, in vitro colony-forming unit (CFU) assays were developed to test the function of single hematopoietic progenitor cells. These assays demonstrated that the survival, proliferation, and differentiation of individual cells vary and depend on cytokines later termed colony-stimulating factors.7-9 Some of these factors allowed for a less restricted lineage output than others (eg, producing both granulocytes and macrophages, rather than granulocytes exclusively), pointing toward a hierarchical structure of hematopoietic progenitors with increasing lineage commitment.
Since then, technological advances have progressively refined this model. The development of flow cytometry–based immunophenotyping made it possible to purify “gated” populations of cells using monoclonal antibodies against cell surface markers.10-13 Certain marker combinations defined hematopoietic cells with different potencies, demonstrating heterogeneity within the hematopoietic stem and progenitor cell (HSPC) compartment.14-17 This led to the phenotypic definition of lineage-committed populations such as common lymphoid progenitors,18 common myeloid progenitors (CMP), granulocyte-macrophage progenitors (GMP),19 lymphoid-primed multipotent progenitors,20 and multilymphoid progenitors.21 The emergence of transcriptomic technologies enabled genome-wide profiling of these purified populations, revealing gene expression changes across stages of hematopoiesis.22-24 Additionally, assessing chromatin accessibility and epigenetic marks such as histone modifications highlighted how epigenetic regulation affects stem cell properties and often precedes transcriptional changes.25-27 More recently, the emergence of high-throughput single-cell methods revolutionized the field, offering unprecedented granularity and resolution within the hematopoietic compartment.28-30 The ability to quantify expression levels of thousands of genes for thousands of individual cells allows for marker-independent identification of cell types (eg, HSCs, GMPs, and T cells) and activity programs (eg, stress, inflammation, and exhaustion), inviting us to refine models of hematopoiesis with enhanced molecular precision and to revisit concepts relating to stem cell differentiation. This review aims to characterize how recent advances in single-cell technologies shape our understanding of hematopoiesis.
Advancements in hematopoiesis through single-cell genomics
Traditionally, single blood cells have been phenotypically profiled using flow cytometry and functionally assessed using CFU and transplantation assays. The development of single-cell genomics over the last decade complements these analyses and allows us to get a glimpse of internal cellular processes, recovering an unprecedented amount of information within a single cell for thousands of cells in an experiment.31
The earliest and most common single-cell sequencing technology is single-cell RNA sequencing (scRNA-seq). Early works relied on quantitative polymerase chain reaction assessing a handful of genes in flow-sorted cells in well arrays.32 Modern scRNA-seq methods rely on next-generation sequencing, assess gene expression levels across the entire genome, and vary in cell separation, with 3 major approaches being well-/plate-based assays (eg, Smart-seq2/3,33,34 MARS-seq,35 and Seq-Well36,37), microfluidics (eg, Drop-seq38 and 10x Genomics Chromium39), and combinatorial indexing (eg, sci-RNA-seq40 and SPLiT-seq41). scRNA-seq has been widely used to characterize the hematopoietic compartment, for example, to characterize HSC heterogeneity in combination with flow sorting and functional assays,42 to reject the simplistic division into CMPs and common lymphoid progenitors,43 and to identify subpopulations of GMPs with variable lineage commitment.44
Gene expression captures only one aspect of cellular heterogeneity. Analyzing different modalities can provide additional insight. For example, changes in epigenetic modifications and chromatin state are pivotal for gene regulation. Both bulk and single-cell assay for transposase-accessible chromatin with high-throughput sequencing (scATAC-seq) have shown that changes in chromatin accessibility precede changes in gene expression during differentiation.25,45 Chromatin accessibility data also provides greater sensitivity in inferring the activity of transcription factors compared with scRNA-seq, owing to the low expression levels and high dropout rates of these proteins. scATAC-seq data revealed decreased accessibility of HOX transcription factor motifs in preleukemic HSCs. Knockdown of HOXA9 in healthy HSPCs increased their stem cell phenotype and decreased their differentiation capacity, validating this observation.46 Histone modifications can provide more specific insights into chromatin structure, revealing active, repressed, or poised states of chromatin and differentiating the activity of certain regulatory elements. Profiling mouse bone marrow cells for 4 histone modifications recently revealed the interplay between euchromatin and heterochromatin dynamics during hematopoiesis.47 DNA methylation, another important layer of epigenetic regulation, provides stable, lineage-specific markers. Hui et al demonstrated that subpopulations within HSCs can robustly be identified using a small number of stochastically sampled CpGs, a significant benefit considering high dropout rates in single-cell experiments.48 Furthermore, DNA methylation can be used to infer lineage histories, as has been demonstrated for chronic lymphocytic leukemia,49 and, more recently, in native hematopoiesis in mice and humans.50
Because of the complexity of posttranscriptional regulation, transcript levels cannot be used as a proxy for protein abundance. Technologies such as CITE-seq51,52 and REAP-seq53 bridge the gap between transcriptional and phenotypic states of the cell, reporting gene expression and surface protein levels for every cell in the experiment. Taking these approaches to the next level, a transcriptomic atlas of human HSPCs with 132 surface markers recently revealed novel myeloid and erythroid lineage markers, and allowed alignment of malignant cell states.52
Combining multiple modalities within the same single-cell experiment (multiomics approach) increases the power to resolve heterogeneity and gain mechanistic insights. Gene expression readout can be currently combined with protein levels,51 chromatin accessibility,54-56 genome mutations,57-59 mitochondrial variants,60 T-cell receptors,61,62 and DNA methylation.63,64 Additionally, several lineage tracing approaches can be combined with scRNA-seq to assess the differentiation routes of individual cells, as reviewed in “Lineage fates and biases.”65 In dendritic cell leukemias, combined analysis of gene expression, DNA mutations, and T-cell receptor sequences provided insight into disease ontogeny and immune dysregulation.66,67 A recent preprint combined gene expression, DNA mutations, chromatin accessibility, and surface protein levels to show increased activity of the prosurvival unfolded protein response component ATF4 in UBA1-mutant cells, validated through ATF4 knockout and UBA1 inhibition experiments in human VEXAS (vacuoles, E1 enzyme, X-linked, autoinflammatory, somatic) syndrome samples.68
Evolving views of hematopoietic differentiation
Understanding the hematopoietic hierarchy and the regulatory circuits that govern quiescence, lineage priming, and differentiation is crucial. Quantitative insight into the complex process of hematopoiesis directly connects to the mechanisms driving blood disorders and the development of targeted therapies.
Increased appreciation of cellular heterogeneity
The activity of gene expression programs associated with quiescence and lineage priming has been described using single-cell genomics.28,44,69-74 A common theme in these studies is the discovery of heterogeneity in cell populations that were previously thought, based on flow cytometry gating, to be relatively homogeneous.28,72,75 This included the discovery of differential HSC dormancy states influenced by retinoic acid signaling, validated by retinoid acid treatment in vitro, and by a vitamin A–free diet in mice76; identifying subpopulations of GMPs with variable lineage commitment and a bipotential poised state coexpressing Irf8 and Gfi1 (antagonistic determinants of macrophage-neutrophil fate decision), which was validated by CFU assays and Irf8/Gfi1 knockouts44; characterizing new subpopulations of dendritic cells validated by flow cytometry and differential responses and cytokine profiles in functional assays.77 These observations prompted a paradigm shift in our understanding of hematopoiesis, moving away from the conventional model portraying neatly demarcated cell types that can be purified to near functional homogeneity.
Transitions: discrete vs continuous
The observed heterogeneity raises the question of the level of “connectivity” between different stem and progenitor populations, or the extent to which transitions between phenotypic states can be described as “discrete.” In the classical model proposed >20 years ago, hematopoiesis is depicted as a branching structure with discrete bifurcations producing uniform populations defined by surface marker expression.78,79 This representation is a useful conceptual tool; however, it is a simplification that does not represent the more gradated nature of HSPC commitment that is evident from both flow cytometry and single-cell sequencing data. This was readily demonstrated with the emergence of scRNA-seq data, with progenitor populations occupying adjacent and overlapping areas in transcriptional space.80,81 Some of the early works suggested that HSPCs exist in an unstructured transcriptional continuum of low-primed cells that produce unilineage-restricted progenitors.80,82 Others reported a continuous but structured landscape of the HSPC compartment.70,81,83 The “punctuated continuum” model was proposed to bring together discrete and continuous models, suggesting the existence of punctuated transitions between some of the functionally distinct heterogeneous populations.84 Improvements in both experimental and computational techniques provide increasingly nuanced views of the differentiation landscape, with a recent paper describing multiple adjacent populations of stem, progenitor, and transitional cells in the hematopoietic compartment using a wealth of CITE-seq data.52 Other sensitive scRNA and/or scATAC-seq assays similarly identified discrete human HSPC populations.85-87 Thus, although positioning cells in a multidimensional space using single-cell sequencing has demonstrated more continuity in hematopoietic differentiation than previously appreciated, the resolution afforded by the latest generation of technologies is revealing discernable structures in the HSPC compartment.
Incorporating labeling to accurately infer transition rates
Although single-cell expression data demonstrate relative continuity within the hematopoietic compartment and inform expression (or epigenetic) changes associated with differentiation trajectories, it remains a static snapshot from which transition rates between the populations cannot be inferred. To study transition rates between populations of cells, time-resolved data are required. Label-propagation studies offer an elegant way to solve this problem. In such studies, a genetic label is introduced into a population of HSCs, which is then propagated at different rates across different parts of the hematopoietic compartment, reflecting self-renewal and differentiation rates of progenitor populations. By sorting predefined populations of cells to quantify their progeny, this approach has highlighted the gradual loss of self-renewal during HSC differentiation and dissected different routes of megakaryopoiesis.71,88 Recently, label propagation was integrated with single-cell transcriptomic data, allowing for self-renewal and differentiation rate assessment across the transcriptional continuum.89 The authors estimated the flux between subpopulations of HSCs and progenitors and inferred expression changes accompanying these transitions. Expression changes demonstrated poor correlation with differentiation rates, implying that transcriptional differences alone cannot be used to derive the real-time differentiation dynamics. Furthermore, transplantation accelerates the differentiation of progenitor cells.89,90 Because this acceleration can reduce the likelihood of state transitions, transplantation assays may underestimate HSPC fate potential (see also “Transplantation vs native hematopoiesis”). Thus, our view of hematopoiesis should incorporate progenitor populations with near self-renewal that limit state transitions in conditions that accelerate differentiation.
Lineage fates and biases
To quantify the potency and lineage bias of single HSPCs, it is necessary to follow the fate of individual cells. Single HSC transplantation has historically been the gold standard for assessing HSC fate and functional heterogeneity.91,92 As a more high-throughput alternative, genetic barcoding techniques have emerged to trace the output of many cells in a single experiment. In a lineage tracing experiment, a population of cells (usually HSCs) is labeled with individual barcodes, whose propagation across different hematopoietic compartments can be measured over time, producing lineage outputs of individual cells.
Artificial barcodes to track the output of individual HSPCs
To characterize the heterogeneity and lineage contributions of HSCs,93 CMPs,94 and lymphoid-primed multipotent progenitors,95 these populations can be transduced with a library of barcode-containing lentiviral constructs followed by fate tracking in vitro or in vivo. In approaches like LARRY (lineage and RNA recovery), barcodes are introduced under a ubiquitous promoter, allowing for the simultaneous inference of the barcode and the transcriptional state of a cell.93,96 This method demonstrated structured continuity of early decisions in the hematopoietic compartment, inferred transcription changes associated with gradual lineage commitment,96 and identified low-output HSCs with the highest quiescence and serial engraftment potential.93 The ability to combine molecular information with functional readout is crucial to identify and validate the underlying regulatory molecules.44,93 A recent preprint delivered lentiviral barcodes into 6 major populations of HSPCs to comprehensively characterize the in vivo fate of 5018 HSPCs and analyze their contributions to mature lineages.97 Their results suggest a model in which hematopoiesis occurs along multiple parallel “tracks,” through which some HSCs undergo early commitment toward certain lineages and stay in their tracks down the differentiation route.
To study lineage contributions in native hematopoiesis, barcodes need to be introduced in a living animal. Such approaches include Cre–induced random recombination events,98 inducible transposon insertion,99,100 and CRISPR-Cas9–based approaches.101,102 Early work in native hematopoiesis showed that mouse HSCs contribute minimally to mature blood cell production, with early progenitors producing most of the progeny instead.99 This likely reflects low differentiation rates of the most primitive HSCs. A notable contribution of HSCs of both unilineage and multilineage potential was reported in a recent study.103 The extent to which HSCs contribute to unperturbed hematopoiesis remains controversial, potentially because of different HSC definitions or a low number of assessed HSCs.88,104 In vivo lineage tracing identified megakaryocyte-restricted HSCs acting in parallel to the conventional megakaryocyte differentiation path,100,105 in agreement with other works.71,82,106 These megakaryocyte-restricted HSCs may acquire multilineage potential under transplantation settings,100 suggesting that HSCs can demonstrate plasticity when prompted by certain conditions.
Transplantation vs native hematopoiesis
This observation illustrates an important issue in hematology research: the behavior of hematopoietic cells in functional assays may differ from native hematopoiesis. Experimental systems often include perturbations such as in vitro expansion or in vivo transplantation. However, these conditions can accelerate HSPC commitment, which may introduce lineage restriction that is not representative of native hematopoiesis.90,107 Transplantation assays appear to accentuate HSC heterogeneity and lineage bias compared with unperturbed hematopoiesis. A study in CARLIN (CRISPR array repair lineage tracing) mice found that cytotoxic myeloablation prompts HSCs to contribute to hematopoiesis more actively and unevenly, with a minority of highly active HSCs showing a high level of proliferation.101 This is reminiscent of molecular reprogramming during emergency myelopoiesis.108 Relatedly, conditioned transplantation led to a biased contribution from dominant HSCs, whereas unconditioned transplantation mirrored unperturbed conditions.109 This concept was supported and extended by label propagation studies demonstrating that transplantation accelerates differentiation, limiting HSPCs from exploring different developmental paths available under native conditions, hence leading to a restriction in the observed fate potential.89 Thus, conditions that stress the system alter HSC differentiation dynamics, which can limit the extent to which experimental systems represent native hematopoiesis.
Leveraging naturally occurring genetic variation
Most observations on the dynamics of hematopoietic lineages come from mouse models. Although these have provided invaluable information, some aspects of mouse hematopoiesis cannot be directly extrapolated to humans.110 Tractable experimental systems to study human hematopoiesis include CFU assays, (humanized) xenograft models, and induced pluripotent stem cells.111,112 Even so, these do not capture the full genetic, environmental, and temporal complexity of native hematopoiesis. A promising supplement for clonal analysis of human hematopoiesis is to use naturally occurring barcodes like genomic mutations, mitochondrial variants, or inheritable epigenetic changes as retrospective lineage markers.65 Naturally occurring somatic mutations in the nuclear genome can be informative lineage markers. By identifying somatic variation in individual HSPCs through a CFU assay followed by whole-genome sequencing and leveraging methods of evolutionary biology, we can infer the size and dynamics of a stem cell compartment, estimate the contribution of HSCs to various lineages, and observe that clonal diversity decreases with age.113,114 However, CFU assays cannot be combined with the detailed cell state measurements that single-cell genomics can provide. To combine scRNA-seq with lineage tracing, large-scale events like copy number alterations can be used to infer clonal evolution within a tumor, but they are rare in healthy tissues and cannot define clonal lineages comprehensively.115 Mitochondrial variants provide a promising source of natural genetic variability, capable of detecting clonal populations within human samples.60,116,117 Recent advances that cover mitochondrial variants at greater depth allowed researchers to characterize the functional heterogeneity of HSCs and confirm age-associated decline in clonal diversity, while concurrently obtaining single-cell gene expression and chromatin accessibility states.87 In the study by Weng et al,87 HSC clonal groups exhibited limited but stable lineage bias, potentially reflecting cell plasticity in native hematopoiesis. Epigenetic changes provide a potential nongenetic source of heritable changes that can be assessed at the single-cell level, as demonstrated in a recent preprint on mice and human native hematopoiesis, which assessed changes in clonal diversity with age,50 and a study of chronic lymphocytic leukemia evolution.49 Native lineage markers have their own technical limitations (eg, mitochondrial variants are heteroplasmic and can be lost because of genetic drift or undergo horizontal transfer). The resolution of these methods can provide is being actively explored.
Perspective
Over time, our views of hematopoiesis have evolved substantially. The classical view of hematopoiesis posits that phenotypically and functionally defined stem and progenitor cells are clearly demarcated into discrete states. The early single-cell studies challenged this view, supporting a continuous differentiation trajectory. Based on the latest data, we now conclude that the structure of the HSPC compartment falls between the previously proposed extremes, representing hematopoiesis as a complex and gradual process with transitory states. Cell populations accumulate in basins created by attractor points scattered across the hematopoietic differentiation landscape (Figure 1A).118 Transitions between attractor states are governed by transcriptional, epigenetic, and environmental changes that elicit differentiation trajectories. Some basins are shallow and less stable, reminiscent of a metastable state.44 Some metastable states are located close to each other on the differentiation landscape allowing for easier transitions between them; flux between states with varying fate potentials forms multipotent/oligopotent progenitor populations. Further down differentiation trajectories, transitions between separate basins become prohibited, requiring a nonphysiologic level of metabolic perturbations, producing unipotent progenitors and their terminally differentiated progeny. Although cell type identity becomes fixed, external stimuli can still alter activity programs such as inflammation and exhaustion. Although remaining technical limitations still complicate a precise description of the punctuated states and transition rates, we believe transcriptional and epigenetic analysis has potential in resolving HSPC states that were previously unknown.
Structure and connectivity of the hematopoietic landscape. (A) Schematic representation shows cell populations with various differentiation potentials on the Waddington landscape. The panels on the right show how different levels with local minima correspond to attractor states and transition likelihoods (indicated by arrow width). (B) The degree of connectivity between HSPC subpopulations is not yet fully understood. Under transplantation settings, lineage commitment is established in some HSCs and maintained in downstream progenitors, forming multiple tracks of differentiation (left). The landscape of steady-state hematopoiesis may allow for more transitions and HSPC fate plasticity (right). MPP, multipotent progenitor.
Structure and connectivity of the hematopoietic landscape. (A) Schematic representation shows cell populations with various differentiation potentials on the Waddington landscape. The panels on the right show how different levels with local minima correspond to attractor states and transition likelihoods (indicated by arrow width). (B) The degree of connectivity between HSPC subpopulations is not yet fully understood. Under transplantation settings, lineage commitment is established in some HSCs and maintained in downstream progenitors, forming multiple tracks of differentiation (left). The landscape of steady-state hematopoiesis may allow for more transitions and HSPC fate plasticity (right). MPP, multipotent progenitor.
A recent study suggests a multitrack model of hematopoiesis.97 It reports the existence of early lineage commitment in some HSCs, which canalizes the entire differentiation route of their progeny, implying transitions between HSCs and early progenitors are deterministic, and horizontal transitions between parallel tracks are restricted. Besides the megakaryocyte-restricted populations of HSCs, the relation of this transplantation-based model to native hematopoiesis is yet unclear (Figure 1B). As discussed above, transplantation and other bone marrow insults reshape the differential landscape to meet the demand of perturbations, accelerating differentiation and lineage commitment. As a result, potential plasticity existing in a cell may remain unrealized. In steady-state hematopoiesis, transitions between the states may be more stochastic, allowing for some flexibility in the system output.89 The future development of unperturbed lineage tracing methods will allow us to sample the entire spectrum of routes hematopoietic cells can take, providing insight into the degree of plasticity of this vital system, and how plasticity is affected by stress and pathological conditions.
We are still in the early stages of single-cell technologies, and they face challenges such as relatively low output and high dropout events. Although combining multiple modalities can mitigate these issues, continuous innovation in experimental protocols remains crucial. The growing popularity of single-cell genomic tools is also accompanied by the development of new bioinformatic approaches to solve problems that single-cell data analysis posed,119 including batch correction,120 integration of data across multiple data sets121-123 and modalities,124,125 cell cluster identification and annotation,126,127 and data visualization.124,125,128,129 Cell atlases are actively being developed alongside efficient annotation transfer methods to facilitate cell cluster annotation and novel disease marker identification.124,125,128,129 Continued efforts to create atlases that encompass novel disease states will enhance our ability to compare altered cells with their healthy counterparts.130,131 Data sets combining transcriptomic information with surface proteins panels will pave the way toward new standards in flow cytometry gating strategies in research, diagnostics, and treatment.52,132 Overall, advancements in single-cell technologies, computational methods, and public bioinformatic resources will continue to provide a more accurate picture of the multiple layers of heterogeneity underlying complex biological systems, including hematopoiesis.
Additionally, the integration of single-cell methods with functional readouts and lineage tracing approaches is expected to deepen. By combining single-cell sequencing with functional genomics, we can test how candidate genes alter the hematopoietic differentiation landscape.133 Genetic labels that are accumulated at a fixed rate have the potential to efficiently sample the ensemble of allowed differentiation trajectories; we expect that systems similar to Cas9–based molecular recorders will move toward enhanced temporal resolution, increased barcoding efficiency, and compatibility with single-cell sequencing.134-136 In parallel, the development of better methods and algorithms to trace naturally occurring barcodes will be necessary to study human samples.
The implementation of single-cell genomics has advanced our understanding of blood cell production with age, clonal hematopoiesis, and blood cancers. Aging-associated changes, including increased myeloid and platelet output, changes in T-cell composition, and decreased clonal diversity are readily identified and refined through single-cell data analysis.50,87,105,113,137,138 Recently, the impact of inflammation on HSCs, the existence of their inflammatory memory, and its connection with aging have been reported.85,139,140 Studying the hematopoietic compartment of centenarians can provide insights into how the immune system maintains its function with age, suggesting targets to prolong immune health.141
Clonal hematopoiesis occurs as HSCs acquire somatic mutations that confer a proliferative advantage, leading to the expansion of clones, as explored in several single-cell studies.86,142,143 However, most mutations that confer a proliferative advantage to HSCs do not lead to malignant transformation, potentially because they bend the hematopoietic landscape differently depending on where they land. Pairing cell state analysis with lineage tracing is beginning to pinpoint clonal lineages of cells that contribute to the progression of clonal hematopoiesis to malignancy, as demonstrated by combining single-cell transcriptomics with lineage tracing of native barcodes to trace leukemia stem cells among their healthy counterparts.144
In malignancies, targeted single-cell genotyping can infer the complex dynamics of clonal hierarchies in malignancies. When applied to clonal hematopoiesis, myeloproliferative neoplasms, acute myeloid leukemia, and acute lymphoblastic leukemia, this approach helps associate specific genes with clonal advantages, evaluate treatment responses, identify residual disease, and target minor clones with relapse potential.145,146 Single-cell genomics, linked with drug response data, can help the design of precise interventions to target cellular heterogeneity underlying drug resistance.147 An example of the potential impact of such an approach is seen in acute myeloid leukemia, in which cell heterogeneity reflects stages of healthy myeloid differentiation and directly affects drug sensitivity and clinical responses.148-151 As we chart the complex landscape of hematopoiesis, both expected and unexpected opportunities to enhance healthy hematopoiesis will unfold.
Acknowledgments
P.v.G. is supported by the National Institutes of Health, National Cancer Institute (grant R33 CA278393), the Edward P. Evans Foundation, the Vera and Joseph Dresner Foundation, the Ludwig Center at Harvard, the Starr Cancer Consortium (grant I15-0015), and the Brigham Research Institute.
Authorship
Contribution: K.S. and P.v.G. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Peter van Galen, Department of Hematology, Brigham and Women’s Hospital, Harvard Institutes of Medicine, 4 Blackfan Circle, Boston, MA 02115; email: pvangalen@bwh.harvard.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal