In this issue of Blood, Zhang et al1 use a combination of bulk and single-cell sequencing analyses of diagnosis and relapse paired samples of patients with T-cell acute lymphoblastic leukemia (T-ALL) to characterize nonleukemic T cells within the leukemia milieu, to expose 2 different patterns of leukemia clonal evolution upon treatment, and to identify the RNA-binding protein Musashi-2 (MSI2) as a key player in T-ALL therapy-induced clonal evolution and resistance to chemotherapy.
There have been remarkable improvements in treatment outcome of childhood T-ALL. However, refractory disease and relapse remain the main clinical challenges in this malignancy. The findings of Zhang and collaborators have obvious clinical potential to help address relapsed or refractory disease. For example, by simultaneously conducting transcriptomic and T-cell receptor (TCR) sequencing in single cells, the authors distinguish T-ALL cells from nonleukemic T cells in the same patient. They find that the frequency of γδ T cells, which can mount immune responses against T-ALL,2 and of CD8 effector and proliferating T cells is higher (whereas that of naive T cells is lower) in patients at diagnosis that responded to therapy than in healthy controls. This is expected if an antitumoral immune response is occurring. However, in diagnostic samples of patients that relapsed, the frequency of CD8 effector and proliferating T cells, as well as of γδ T cells, is lower, resembling healthy controls. Moreover, the proportion of infiltrating T cells with the same TCR (indicative of specific T-cell activation) at diagnosis is higher in nonrelapsed patients than in relapsed patients. Notably, exhausted CD8 T cells increase in frequency from diagnosis to relapse. These findings suggest that the immune system plays an important role in the response to chemotherapy in T-ALL and raise the tantalizing possibility that the composition of immune cell infiltrates at diagnosis may serve as a marker for those patients more likely to respond to therapy. The potential clinical implications of these observations, including exploring rational combinations of chemo- and immunotherapy, are considerable.
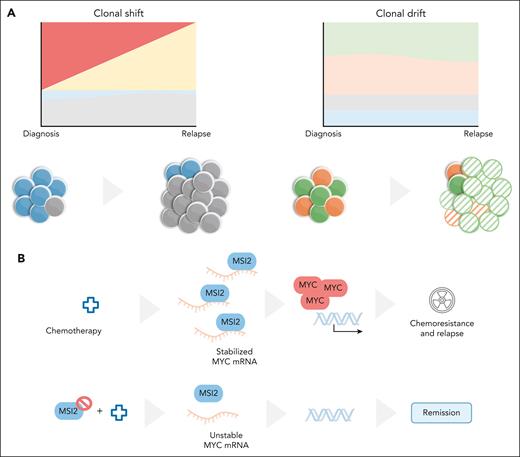
Darwinian evolution applies to cancer cells, including, evidently, during exposure to treatment. In agreement with recent studies by Alberti-Servera and collaborators,3 Zhang et al show that, in some patients, there are obvious alterations in clonal composition from diagnosis to relapse, with some subclones progressively enriching throughout the course of chemotherapy. This evolutionary pattern, which the authors refer to as “clonal shift,” has been reported in T-ALL and many other cancers.3,4 However, another pattern, coined “clonal drift,” was described by Zhang et al for T-ALL, in which there is no substantial shift in clonal composition but there are considerable changes in the transcriptional profile at relapse (see figure). Thus, 2 alternative evolutionary patterns occur in response to therapy that are characterized by either “dynamic” (clonal shift) or “stable” leukemic clones, with the variation occurring instead in the transcriptome (clonal drift). Fitness in this case likely reflects selection of traits that are epigenetically determined. In other words, “cell states” rather than “cell clones” are the subject of Darwinian selection—similarly to what Turati et al have recently reported for childhood B-cell acute lymphoblastic leukemia (B-ALL).5
T-ALL clonal dynamics and MSI2. (A) Two patterns of clonal evolution in response to treatment were identified in T-ALL. In clonal shift, there is substantial reshuffling of clonal composition after treatment, whereas in clonal drift no major clonal alterations occur, with most variation from diagnosis to relapse occurring at the transcriptional level. (B) Analysis of clonal drift resulted in the identification of a MSI2-MYC axis driving T-ALL relapse that can be counteracted by small molecule inhibition of MSI2. mRNA, messenger RNA. Illustration by Marta B. Fernandes.
T-ALL clonal dynamics and MSI2. (A) Two patterns of clonal evolution in response to treatment were identified in T-ALL. In clonal shift, there is substantial reshuffling of clonal composition after treatment, whereas in clonal drift no major clonal alterations occur, with most variation from diagnosis to relapse occurring at the transcriptional level. (B) Analysis of clonal drift resulted in the identification of a MSI2-MYC axis driving T-ALL relapse that can be counteracted by small molecule inhibition of MSI2. mRNA, messenger RNA. Illustration by Marta B. Fernandes.
What are the determinants of selection resulting in resistance in clonal drift? Commonly enriched “drifted” gene signatures show upregulation of genes such as NFKBIA, SERPINB1, CD69, and MSI2 at relapse. The authors focus on MSI2, a logical choice considering that MSI2 has been associated with poor outcome in ALL.6,7 Now, 2 independent T-ALL cohorts provide evidence that high MSI2 expression at diagnosis is associated with persistence of residual leukemia after induction chemotherapy. This hints at the possibility that MSI2 may be a biomarker of resistance in T-ALL.
So, what leads to MSI2 upregulation? Zhang and colleagues do not provide a definitive answer, although they do show that histone marks (H3K4me3 and H3K27ac) compatible with increased transcription initiation of MSI2 are elevated upon relapse. The answer to the obvious next question is perhaps more relevant: how does MSI2 promote resistance to therapy? Zhang et al show that MSI2 binds to transcripts of the oncogene MYC, thereby stabilizing them and contributing to T-ALL cell viability and proliferation. In vivo evidence in a mouse model of activated NOTCH1-induced T-ALL provides correlative evidence that downregulating MYC by pharmacologically inhibiting MSI2 may be a valid frontline strategy to treat T-ALL. Important for relapse or refractory disease is the demonstration that MSI2 overexpression induces in vitro resistance to daunorubicin, cytarabine, vincristine, and methotrexate, whereas knocking out MSI2 sensitizes T-ALL cells to chemotherapy—an effect that is counterbalanced by MYC overexpression. The corollary is that MYC upregulation is critical for MSI2-mediated chemoresistance in T-ALL. In animal models, leukemia stem/initiating cells display high MSI2 levels,8 T-ALL leukemia initiating cells express high MYC levels, and pharmacological inhibition of MYC activity leads to T-ALL remission.9 Conversely, B-ALL cells that escape chemotherapy are quiescent, with downregulation of MYC activation being a frequent feature of relapsed B-ALL.5 Hence, the MSI2-MYC axis, which can drive resistance in T-ALL, is unlikely to play a role in B-ALL relapse.
Although many questions arise from the work by Zhang et al, such as whether MSI2 expression in T-ALL cells may drive resistance also by impacting the normal immune cell compartment, one point of special interest is the demonstration that small molecule inhibition of MSI2 sensitizes T-ALL cells to chemotherapy in vitro and substantially delays leukemia progression in vivo in patient-derived xenograft models when given in combination with daunorubicin or cytarabine. Although other combinations, including with glucocorticoids, still require analysis, these experiments highlight the potential of MSI2 as a target for therapeutic intervention in relapse/refractory T-ALL.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal