In this issue of Blood, Vallée and colleagues have distinguished 2 classes of WAS variants in patients with Wiskott-Aldrich syndrome (WAS, Online Mendelian Inheritance of Man #301000), which provide, for the first time, a molecular biomarker of disease severity and outcome.1
WAS was first described in 1937 in Germany by Wiskott2 and later in the United States by Aldrich et al3 as a clinical entity comprised of thrombocytopenia, eczema, recurrent infections, autoimmunity, and cancer risk. Its presence in male children predicted the X-linked inheritance that was confirmed on WAS gene cloning in 1995.4WAS encodes a protein called WASP exclusively in hematopoietic cells, which is critical for actin assembly and lymphocyte function.5 WAS is caused by loss-of-function and gain-of-function mutations in WASP that disrupt the actin cytoskeleton on which lymphocytes are particularly dependent for cell motility and cell-cell interaction functions. WAS is the prototypic inborn error of immunity disease, often treated by hematopoietic stem cell transplant or gene therapy.6
Despite decades of work on WAS, the field lacked any biomarker to predict disease severity or outcomes for individuals from the broad spectrum of observed clinical phenotypes. The work presented by Vallée and colleagues in this issue closes this gap for the first time by studying 577 patients with a clinical diagnosis of WAS born between 1932 and 2014, followed in 63 centers in 26 countries over 6118 reported patient-years. The authors applied the rigorous variant curation criteria of the American College of Medical Genetics and Genomics/Association for Molecular Pathology7 to the discernible WAS variants seen in 525 individuals. Of these, 55.2% were deemed pathogenic (P), 32.4% likely pathogenic (LP), 11.4% variants of uncertain significance (VUS), and 1% likely benign or benign. Forty of these DNA variants have never been deposited in ClinVar or published, so this cohort identifies a significant number of novel WAS variants, and the curations provided by Vallée et al in the supplemental table are an important contribution to variant curation consistency for the field.
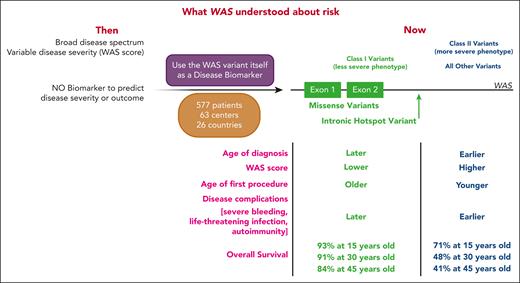
From individuals with P, LP, or VUS WAS variants, the authors looked for genotype-phenotype correlations (see figure). They delineate 2 classes of DNA variants with distinct disease severity and outcome. Class I variants, conferring less severe disease, consist of missense variants encoded by exons 1 or 2 as well as the splice site variant c.559+5G>A, all of which result in lower WASP levels. All other WAS variants were grouped into class II. By every measurement of disease phenotype, individuals with class I variants have less severe disease: later age of diagnosis; lower WAS score; older age of first procedure; lower incidence of severe events with age; later occurring severe bleeding episodes, severe infections, and autoimmunity; and better overall survival, which holds even when only people with P or LP variants are examined (see Figure 4B in the article by Vallée et al).
Schematic representation of WAS disease risk prior to vs after Vallée et al. (Left) Depiction of what was known about WAS disease risk prior to the current publication. WAS comprised a broad spectrum of disease states, and disease severity was estimated using the WAS score. There were no biomarkers to predict disease severity or outcome. (Middle) In this publication, the authors used WAS variants themselves as disease biomarkers, defining class I vs class II variants based on a large cohort of 577 patients with WAS-consistent phenotypes from 63 centers in 26 countries. (Right) Class I variants (green) were comprised of missense variants in exons 1 or 2, or the hot spot intronic variant c.559+5G>A. Individuals with these variants had a less severe phenotype, characterized by later age of diagnosis, lower WAS score, older age of first procedure, later onset of disease complications, and better overall survival compared with all other variants, which were grouped as class II. Created with BioRender.com.
Schematic representation of WAS disease risk prior to vs after Vallée et al. (Left) Depiction of what was known about WAS disease risk prior to the current publication. WAS comprised a broad spectrum of disease states, and disease severity was estimated using the WAS score. There were no biomarkers to predict disease severity or outcome. (Middle) In this publication, the authors used WAS variants themselves as disease biomarkers, defining class I vs class II variants based on a large cohort of 577 patients with WAS-consistent phenotypes from 63 centers in 26 countries. (Right) Class I variants (green) were comprised of missense variants in exons 1 or 2, or the hot spot intronic variant c.559+5G>A. Individuals with these variants had a less severe phenotype, characterized by later age of diagnosis, lower WAS score, older age of first procedure, later onset of disease complications, and better overall survival compared with all other variants, which were grouped as class II. Created with BioRender.com.
Although individuals with class I WAS variants have less severe disease, the authors emphasize that this description is relative to those with an even worse phenotype. All patients with WAS and their family members and caregivers need personalized assessments. The work presented here allows health care providers to give data-based predictions of disease severity, which should help patients and their support systems to develop comprehensive care plans for them. The authors also highlight that the WAS score is intended to be a static measurement of disease severity at one point in an individual’s disease trajectory and is not meant to provide predictive information. Thus, the genotype-phenotype relationships delineated in this study are an important advance for the field. Finally, the authors are to be commended for working together in an international consortium that provided data never before assembled for a rare disease and is a model for how others can collaborate productively to move a field forward.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal