Circulating multilymphoid progenitors are primed for the seeding the thymus and contribute to steady-state T-cell lymphopoiesis.
cHSPCs are responsible for clonal redistribution among BM niches and actively participate in human hematopoiesis.
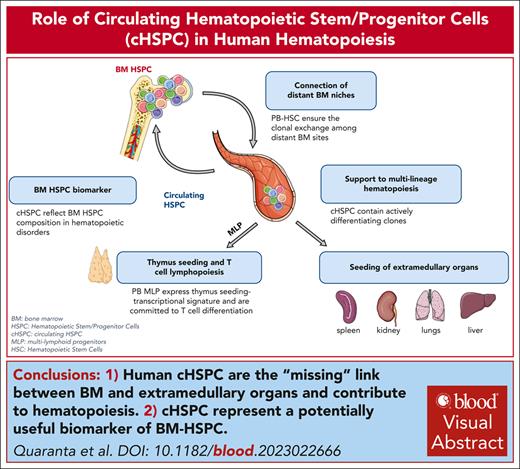
Visual Abstract
In physiological conditions, few circulating hematopoietic stem/progenitor cells (cHSPCs) are present in the peripheral blood, but their contribution to human hematopoiesis remain unsolved. By integrating advanced immunophenotyping, single-cell transcriptional and functional profiling, and integration site (IS) clonal tracking, we unveiled the biological properties and the transcriptional features of human cHSPC subpopulations in relationship to their bone marrow (BM) counterpart. We found that cHSPCs reduced in cell count over aging and are enriched for primitive, lymphoid, and erythroid subpopulations, showing preactivated transcriptional and functional state. Moreover, cHSPCs have low expression of multiple BM-retention molecules but maintain their homing potential after xenotransplantation. By generating a comprehensive human organ-resident HSPC data set based on single-cell RNA sequencing data, we detected organ-specific seeding properties of the distinct trafficking HSPC subpopulations. Notably, circulating multi-lymphoid progenitors are primed for seeding the thymus and actively contribute to T-cell production. Human clonal tracking data from patients receiving gene therapy (GT) also showed that cHSPCs connect distant BM niches and participate in steady-state hematopoietic production, with primitive cHSPCs having the highest recirculation capability to travel in and out of the BM. Finally, in case of hematopoietic impairment, cHSPCs composition reflects the BM-HSPC content and might represent a biomarker of the BM state for clinical and research purposes. Overall, our comprehensive work unveiled fundamental insights into the in vivo dynamics of human HSPC trafficking and its role in sustaining hematopoietic homeostasis. GT patients’ clinical trials were registered at ClinicalTrials.gov (NCT01515462 and NCT03837483) and EudraCT (2009-017346-32 and 2018-003842-18).
Introduction
Hematopoietic stem/progenitor cells (HSPCs) are responsible for the continuous production of short-living mature blood cells. Adult hematopoiesis takes place in the bone marrow (BM), however, rare HSPCs can be also found in the peripheral blood (PB).
In mice, circulating HSPCs (cHSPCs) play an immune-surveillance role by producing tissue-resident innate immune cells, both at steady-state and during tissue-localized infections.1 In case of systemic infections, cHSPCs are mobilized to the spleen to replenish myeloid immune cells.2 Few and fragmented investigations have been conducted so far to characterize human cHSPCs due to technical difficulties in studying this rare population. These studies showed that cHSPC frequency inversely correlates with several factors including age and sex,3 whereas changes in cHSPC count were described in patients with sepsis4,5 or other pathological conditions.6-9 Notably, we have recently shown the persistence of an “inflammatory” epigenetic signature in cHSPCs after COVID-19 infection and recovery.10 However, no study has evaluated so far the cHSPC subset composition in healthy and diseased hematopoiesis, and the direct relationship between BM-HSPCs and the circulating fraction in humans remains to be investigated.
Similar to mice,11-13 human HSPCs were also found within organs,14 including spleen,15 liver,16,17 and intestine.18 By analyzing the transcriptional profiles of hematopoietic stem cells (HSCs)/multipotent progenitors (MPPs) derived from matched BM, PB, and spleen of deceased healthy donors (HD), a recent work reported that PB-HSCs/MPPs shared higher transcriptional similarity with splenic-HSPC than BM counterpart and were enriched for a CD71+ population with low self-renewal properties and high erythroid lineage commitment, which was maintained in older donors and reduced in patients with defective erythropoiesis.15 These data suggest that cHSPCs might act as a link between BM and tissue-resident-HSPCs, providing continuous replenishment of organ-specific HSPC reservoir, but the identity of the HSPC subsets capable of seeding other hematopoietic and nonhematopoietic peripheral organs remains undefined. Intriguingly, donor-derived HSPCs present in intestine, liver, and kidney allografts can contribute to the long-lasting hematopoietic chimerism and the de novo generation of recipient-tolerant T cells, giving proof of their BM and thymic engraftment capability,16-19 leaving open the possibility that cHSPCs might contribute to hematopoietic homeostasis.
A fine combination of chemokines, growth factors, and adhesion molecules is required to maintain HSPC homeostasis, regulating their retention, recruitment, and maintenance in the BM.20-25 However, most of these factors were identified focusing on the BM retention of HSPCs rather than studying whether these pathways can also modulate HSPC physiological egress with similar efficiency on all HSPC subsets.
Here we combined state-of-the-art multiparametric flow cytometry, cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq), ad hoc designed in vitro and in vivo assays, and integration site (IS) analyses to investigate the phenotypic composition, the transcriptional features, the functional properties, and the in vivo dynamics of human cHSPC subpopulations. To our knowledge, this work represents one of the most comprehensive studies on cHSPCs, unveiling fundamental insights into their contribution to human hematopoiesis.
Methods
Characteristics of donors and patients involved in the study
All HD and patient samples were analyzed after signing written informed consent approved by the ethics committees of IRCCS Ospedale San Raffaele (Protocol_TIGET06 for patients treated with Wiskott-Aldrich syndrome [WAS]-gene therapy [GT] and Protocol_TIGET09 for HD and patients with genetic diseases) and of Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico (protocol_number:_239.2018 for neonatal PB sampling). Pediatric patients and donors’ written consent was signed by their parents.
To characterize cHSPC phenotypic composition and content over aging, PB and BM samples were collected from 110 and 29 alive HDs of different ages, respectively (supplemental Tables 1 and 2, available on the Blood website). For the neonatal group, we analyzed the PB and not the cord blood. To investigate cHSPC as a biomarker of BM-HSPC, we analyzed BM and/or PB samples of 35 pediatric patients with impaired hematopoiesis, including WAS, adenosine deaminase deficiency with severe combined immunodeficiency (ADA-SCID), thalassemia (THAL), and BM failure syndromes (BMFSs), as well as 23 individuals without any hematological alterations, including patients with metachromatic leukodystrophy and mucopolysaccharidosis type 1-Hurler and pediatric and adult HDs (supplemental Tables 3 and 4). No patient received HSPC transplantation or GT treatments.
To retrieve ISs from patients with GT, we collected PB and BM samples from 8 patients with WAS HSPC-GT, treated under early access program, or enrolled in clinical trials (clinical trial identifiers: NCT01515462 and NCT03837483) and European Union Drug Regulating Authorities Clinical Trials Database (numbers 2009-017346-32 and 2018-003842-18). This work is a research-based study, not intended to report on the outcome of these clinical trials and early access programs.
Flow cytometry analyses
BM and PB samples collected from HDs, patients with GT, and mice who underwent xenotransplant were analyzed using whole blood dissection protocol,26 labeling samples with fluorescent antibodies against CD3, CD56, CD14, CD41/61, CXCR4, CD34, CD45RA (BioLegend) and CD33, CD66b, CD38, CD45, CD90, CD10, CD11c, CD19, CD7, and CD71 (BD Biosciences). Samples from xenotransplants were preincubated with a mouse FcR blocking reagent (dilution 1:100, BD Biosciences). Absolute cell quantification was performed by adding precision count beads (BioLegend) to samples before whole blood dissection procedure. Human hematopoietic subsets identified are reported in supplemental Table 5.
All stained samples were acquired through BD Symphony A5 cytofluorimeter (BD Biosciences) after Rainbow beads (Spherotech) calibration. Raw data were collected through DIVA software (BD Biosciences) and analyzed with FlowJo software v10.5.3 (BD Biosciences). Technically validated results were always included in the analyses, and we did not apply any exclusion criteria for outliers.
CITE-seq
Live Bulk (LIN–CD34+), LIN–CD34+CD38–, and LIN–CD34+CD38+ HSPC fractions were fluorescence-activated cell sorting (FACS)-sorted from 5 BM and 5 PB from adult HDs (aged, 18-60 years; supplemental Table 6). Cells were loaded into Chromium 10X Single Cell 3’ Gene Expression chips, and library preparation was performed according to the manufacturer’s specifications (Chromium 10× 3’ and TotalSeq™-A protocols). Detailed sample and library preparation and bioinformatics analyses are described in supplemental Methods.
Statistical analyses
Statistical tests, P values, and r coefficient are specified within each figure graph and/or legend. To test the statistical significance between 2 groups, the Mann-Whitney was used, whereas for comparing >2 groups, Dunn multiple comparison was used after nonparametric Kruskal-Wallis statistic. Correlations were assessed through Spearman r test. Statistical tests were performed using Prism v10.0.0 software (GraphPad). Statistical tests for CITE-seq and IS data analyses are described in the corresponding methods section.
Mouse studies were conducted according to protocols approved by the San Raffaele Scientific Institute and Institutional Animal Care and Use Committee (IACUC 1183).
Results
Quantification of human HSPC subpopulations in healthy and diseased hematopoiesis
To quantify cHSPC subsets in human physiological and diseased hematopoiesis, we analyzed PB samples from 110 HDs of different ages (0-87 years; supplemental Table 1) and 35 pediatric patients with impaired hematopoiesis including WAS, a severe combined immunodeficiency with thrombocytopenia,27,28 ADA-SCID,26,29 THAL, with inefficient erythropoiesis,30 as well as BMFS, showing progressive reduction of BM-HSPC content (supplemental Table 3).31 HSPC subsets were identified according to the gating strategy reported in supplemental Table 5.
We found a progressive reduction of cHSPC count and changes in cHSPC composition throughout aging, with a higher frequency of HSCs and MPPs in newborns and an increase of erythroid progenitors (EPs) in adults (Figure 1A-C-C; supplemental Figure 1A). Most HSPC subtypes were quantitatively increased in young infants (supplemental Figure 1B; supplemental Table 7). By comparing patients’ data with age-matched pediatric HDs, we found a higher cHSPC amount in patients with WAS, in line with the murine disease models,32 and a decreased cHSPC number in patients with ADA-SCID, mirroring their reduced CD34+CD38– compartment in the BM.26 cHSPCs were almost absent in patients with BMFS (Figure 1D). We observed a higher variability among patients with the same disease background than pediatric HDs. This finding might be explained by the wide age ranges in distinct patient groups (Table 3) and not by the cHSPC count fluctuation due to the circadian rhythm, because all PB samplings were performed in the first hours of the morning. Moreover, we found consistency between cHSPC composition and the underlying disease, with a lower proportion of megakaryocyte/EPs (MEP) and megakaryocyte progenitors in patients with WAS and of erythroid–committed progenitors in those with THAL (Figure 1E; supplemental Table 8).
cHSPC count and composition change in association with aging and hematopoietic impairment. (A) Correlation between cHSPC absolute count and age of healthy donors (HDs) analyzed. (B-C) Stacked bar graphs displaying the absolute counts (B) and relative composition (C) of PB-HSPC subsets in HDs of distinct age classes. HDs were classified according to age into neonates (NEON), infants (with 3 subgroups: PED 1, PED 2, PED 3), adolescent (PED 4), adult (AD), and older (AGED), as reported in supplemental Table 1. (D) Cell count of cHSPC in PED1/2 group, PED3/4 group, and patients affected by WAS (n = 12), ADA (n = 11), THAL (n = 7), and BMFS (n = 5). Pediatric individuals are represented as triangle (PED1), circles (PED2), square (PED3), and diamonds (PED4) according to their age. Mann-Whitney statistical test was applied for groups’ comparison, and single P values are reported within the graph. (E) Stacked bar graphs displaying the relative composition of cHSPC subsets in PED1/2 groups, PED3/4 groups, and patients with WAS, ADA and THAL. Results of Mann-Whitney statistical test applied for groups’ comparison are reported in supplemental Table 8. (F) Correlation between the number of colony-forming units (CFUs) obtained from 250 μL of whole PB and the age of healthy donors. (G) Correlation between the CFUs retrieved from 250 μL of whole PB and the absolute counts of cHSPCs in patients with WAS, ADA-SCID, THAL, and BMFS. (H) Stacked bar graph displaying the compositions of BM- and PB-HPSCs derived from age-matched healthy individuals (PED and AD). (I) Circulation indexes (CIs) of the single HSPC subpopulations in pediatric and adult donors. Mann-Whitney statistical test was applied for pediatric vs adult CI comparisons, and single P values are reported within the graphs. Kruskal-Wallis test with Dunn multiple comparisons was applied for CI comparisons among the distinct HSPC subsets, and the results are reported in supplemental Table 9. (B-C,E,H) Data are shown as mean with standard error of the mean (SEM). (I) Each dot represents a single individual, and the colored bars show the median value. (A,F-G) Statistical test for correlation: Spearman r. Spearman correlation coefficient (r) and P values are reported in each figure.
cHSPC count and composition change in association with aging and hematopoietic impairment. (A) Correlation between cHSPC absolute count and age of healthy donors (HDs) analyzed. (B-C) Stacked bar graphs displaying the absolute counts (B) and relative composition (C) of PB-HSPC subsets in HDs of distinct age classes. HDs were classified according to age into neonates (NEON), infants (with 3 subgroups: PED 1, PED 2, PED 3), adolescent (PED 4), adult (AD), and older (AGED), as reported in supplemental Table 1. (D) Cell count of cHSPC in PED1/2 group, PED3/4 group, and patients affected by WAS (n = 12), ADA (n = 11), THAL (n = 7), and BMFS (n = 5). Pediatric individuals are represented as triangle (PED1), circles (PED2), square (PED3), and diamonds (PED4) according to their age. Mann-Whitney statistical test was applied for groups’ comparison, and single P values are reported within the graph. (E) Stacked bar graphs displaying the relative composition of cHSPC subsets in PED1/2 groups, PED3/4 groups, and patients with WAS, ADA and THAL. Results of Mann-Whitney statistical test applied for groups’ comparison are reported in supplemental Table 8. (F) Correlation between the number of colony-forming units (CFUs) obtained from 250 μL of whole PB and the age of healthy donors. (G) Correlation between the CFUs retrieved from 250 μL of whole PB and the absolute counts of cHSPCs in patients with WAS, ADA-SCID, THAL, and BMFS. (H) Stacked bar graph displaying the compositions of BM- and PB-HPSCs derived from age-matched healthy individuals (PED and AD). (I) Circulation indexes (CIs) of the single HSPC subpopulations in pediatric and adult donors. Mann-Whitney statistical test was applied for pediatric vs adult CI comparisons, and single P values are reported within the graphs. Kruskal-Wallis test with Dunn multiple comparisons was applied for CI comparisons among the distinct HSPC subsets, and the results are reported in supplemental Table 9. (B-C,E,H) Data are shown as mean with standard error of the mean (SEM). (I) Each dot represents a single individual, and the colored bars show the median value. (A,F-G) Statistical test for correlation: Spearman r. Spearman correlation coefficient (r) and P values are reported in each figure.
Finally, the number of PB colonies correlated with cHSPC count both in HDs and patients (Figure 1F-G; supplemental Figure 1C-D), indicating that PB-HSPCs are endowed with clonogenic potential.
Altogether, we found that the amount of cHSPC subsets changes during aging, with pediatric individuals displaying higher circulation of HSCs/MPPs, and it is altered in case of hematopoietic impairment.
Human BM-resident and trafficking HSPCs display distinct phenotypic composition
To investigate the relationship between BM-HSPCs and cHSPCs, we compared their composition in age-matched PB/BM of HDs and in PB/BM samples collected from the same individuals, including both HDs and patients with WAS, ADA-SCID, metachromatic leukodystrophy, and mucopolysaccharidosis type 1-Hurler, with the latter 2 presenting no hematological alterations (supplemental Tables 2, 3 and 4). In both adult and pediatric donors and patients, we observed a reduced frequency of primitive HSCs, but an increased fraction of MPPs and multilymphoid progenitors (MLPs) in PB- vs BM-HSPCs (Figure 1H; supplemental Figure 2A). Moreover, we found no correlation between PB and BM composition in individuals without hematopoietic defects, suggesting distinct roles of resident and trafficking HSPCs in hematopoietic homeostasis. On the contrary, the frequencies of PB and BM lymphoid– and megakaryocyte–committed progenitors positively correlated in patients with ADA-SCID and WAS (supplemental Figure 2B-C).
To estimate the trafficking propensity of the distinct HSPC subsets, we calculated the circulation index (CI) as the percentage of BM cells circulating in PB. MLPs had the highest CI among all HSPC subsets, whereas HSC-CI was increased in the pediatric groups (Figure 1I-K; supplemental Figure 2D; supplemental Tables 9-10).
Altogether, we found that resident and circulating HSPCs display distinct composition, implying their diverse biological functions. Moreover, in the presence of severe hematopoietic defects, alterations in BM-HSPC composition are also found in cHSPCs.
cHSPCs display a preactivated state with increased differentiation in vitro compared with their BM counterpart
To compare circulating vs resident-HSPCs in healthy donors, we combined single-cell transcriptome and immunophenotypic analysis by performing CITE-seq33 with antibody-derived tag (ADT)–labeled antibodies recognizing HSPC markers. We sequenced 28 315 BM- and 34 443 PB-HSPCs derived from 5 distinct donors/sources (supplemental Figure A-C).
Unsupervised clustering identified 18 clusters (Figure 2A-B) that were classified as immature (c1, c12, c11, c9, c8, c0), myeloid (c5, transitioning-common myeloid progenitors/granulocyte-monocyte progenitors (CMPs/GMPs); c7, granulocytes progenitors; c10, mono/dendritic progenitors, MDPs), lymphoid (c4, MLPs; c15, Precursor of B/NK cells, PreBNKs; c17, Early T-cell progenitors ETPs, T cells), megakaryocytic (c16), and erythroid (c2, c3, c14, immature-erythroid; c6, c13, mature-erythroid) clusters (Figure 2B; supplemental Figure 4A). Differential expression analysis among immature clusters allowed for the identification of long-term (LT)-HSCs (c1), cycling-HSCs (c12), MPPs (c9), erythroid-MPPs (c8), and cycling-MPPs (c11) (Figure 2C; supplemental Figure 4B). Cluster 0 was partitioned into transitioning MPPs/CMPs (c0.0), erythroid-CMPs (c0.1), myeloid/lymphoid-CMPs (c0.2), and myeloid-CMPs (c0.3) (Figure 2C; supplemental Figure 4C).
Single-cell transcriptional and phenotypic profiling of human PB– and BM–derived HSPC subpopulations. (A) UMAP embedding, coloring cells by BM (red) and PB (blue) sources. (B) UMAP embedding showing 18 seurat clusters identified after unsupervised clustering. (C) UMAP embedding showing seurat clusters 1 to 17 and subclusters of seurat cluster 0. The legend on the right shows the annotation of the single clusters. (D) Stacked bar graph showing the distribution of the transcriptional clusters in BM- and PB-HSPCs. The dominant transcriptional signatures identified for PB-HSPCs are reported. (E) UMAP embedding showing the distinct phenotypic HSPC subpopulations identified by antibody derived tag (ADT) protein barcoding. (F) Distribution of the transcriptional clusters in the 10 ADT–defined HSPC subpopulations. Clusters are grouped according to the expressed HSPC transcriptional signature.
Single-cell transcriptional and phenotypic profiling of human PB– and BM–derived HSPC subpopulations. (A) UMAP embedding, coloring cells by BM (red) and PB (blue) sources. (B) UMAP embedding showing 18 seurat clusters identified after unsupervised clustering. (C) UMAP embedding showing seurat clusters 1 to 17 and subclusters of seurat cluster 0. The legend on the right shows the annotation of the single clusters. (D) Stacked bar graph showing the distribution of the transcriptional clusters in BM- and PB-HSPCs. The dominant transcriptional signatures identified for PB-HSPCs are reported. (E) UMAP embedding showing the distinct phenotypic HSPC subpopulations identified by antibody derived tag (ADT) protein barcoding. (F) Distribution of the transcriptional clusters in the 10 ADT–defined HSPC subpopulations. Clusters are grouped according to the expressed HSPC transcriptional signature.
cHSPCs were mainly categorized in clusters with immature, erythroid, and MLP transcriptional signatures, whereas BM-HSPCs displayed a more balanced distribution across all clusters (Figure 2D; supplemental Figure 4D). Additionally, we found that c9, c11, and c12 were almost entirely composed of BM cells (supplemental Table 11).
By applying our FACS gating strategy on ADT-sequenced cells (supplemental Table 6), we identified all HSPC subpopulations (Figure 2E; supplemental Figure 5A-D;supplemental Table 12). Of note, ADT-defined groups were enriched in the corresponding transcriptional clusters, either expressing primitive or differentiation-primed signatures, thus implying a consistency between HSPC phenotypic and transcriptional lineage commitment (Figure 2F).
To compare transcriptionally consistent populations from PB or BM, we performed intracluster comparisons through differential gene expression followed by gene set enrichment analysis. Given the high redundancy of gene ontology (GO) terms in distinct intracluster comparisons, we applied a semantic reduction algorithm34 to define GO macrocategories associated with PB or BM cells (supplemental Figure 6A-B). PB-HSPCs were enriched in macrocategories associated with differentiation, adhesion molecules, and activation, whereas BM-HSPCs macrocategories were related to high transcriptional, metabolic, and replicative cell states (Figure 3A). Additionally, BM LT-HSCs displayed higher expression of genes related to increased cellular activity, whereas PB LT-HSCs showed upregulation of genes associated with interferon signaling and low transcriptional/proliferative state (Figure 3B). Consistently, we found a higher fraction of S/G2M cells in BM-HSPCs, whereas cHSPCs were predominantly in G1 phase, suggesting their preactivated state. Notably, cells in G0 state, characterized by a “dormancy” transcriptional signature,35-40 were mostly found in BM-resident primitive compartments (Figure 3C-D; supplemental Figure 7A-B).
cHSPCs show a preactivated state associated with higher in vitro differentiation efficiency than BM-HSPCs. (A) Tile plot of the top 5 GO-BP macrocategories for each cluster. Statistically significant GO-BP ontology gene sets (adjusted P < .05) with an enriched expression in PB (NES > 0) or BM (NES < 0) are shown in blue and red, respectively. For each macrocategory, color intensity is proportional to NES absolute values. Macrocategories were classified in diverse groups, according to the associated biological functions. (B) Heat map showing the scaled expression of cluster 1 marker genes that are differentially expressed between PB and BM (adjusted P < .05; logFC ≥0.4 or ≤0.4.) cells. Genes are grouped by biological functions. Annotation for BM (red) and PB (blue) cells is reported. (C) UMAP embedding grouping cells by source and coloring by inferred cell cycle. (D) Histograms representing the percentage of cells in G0, G1, S, and G2M cell cycle phases for each cluster in the BM (top) and the PB (bottom) data sets. The x-axis shows cluster annotation according to the HSPC subset transcriptional signatures. Classification of the cell cycle activity was performed on the overall data set, shown in supplemental Figure 7B, and then reported on BM (top) and PB (bottom) single data sets. (E) Summary of the differentiation efficiency of PB-HSPCs (n = 4) and BM-HSPCs (n = 3) in our single-cell in vitro differentiation assay. The number of seeded wells, the count of wells showing differentiated progeny after 3 weeks of culture, and the total differentiation efficiency starting from one single cell are reported. (F) Pie charts representing the frequencies of BM- and PB-HSPC showing specific lineage scores detected at the end of the single-cell in vitro differentiation assay. GO-BP, gene ontology-biological processes; NES, normalized enrichment score.
cHSPCs show a preactivated state associated with higher in vitro differentiation efficiency than BM-HSPCs. (A) Tile plot of the top 5 GO-BP macrocategories for each cluster. Statistically significant GO-BP ontology gene sets (adjusted P < .05) with an enriched expression in PB (NES > 0) or BM (NES < 0) are shown in blue and red, respectively. For each macrocategory, color intensity is proportional to NES absolute values. Macrocategories were classified in diverse groups, according to the associated biological functions. (B) Heat map showing the scaled expression of cluster 1 marker genes that are differentially expressed between PB and BM (adjusted P < .05; logFC ≥0.4 or ≤0.4.) cells. Genes are grouped by biological functions. Annotation for BM (red) and PB (blue) cells is reported. (C) UMAP embedding grouping cells by source and coloring by inferred cell cycle. (D) Histograms representing the percentage of cells in G0, G1, S, and G2M cell cycle phases for each cluster in the BM (top) and the PB (bottom) data sets. The x-axis shows cluster annotation according to the HSPC subset transcriptional signatures. Classification of the cell cycle activity was performed on the overall data set, shown in supplemental Figure 7B, and then reported on BM (top) and PB (bottom) single data sets. (E) Summary of the differentiation efficiency of PB-HSPCs (n = 4) and BM-HSPCs (n = 3) in our single-cell in vitro differentiation assay. The number of seeded wells, the count of wells showing differentiated progeny after 3 weeks of culture, and the total differentiation efficiency starting from one single cell are reported. (F) Pie charts representing the frequencies of BM- and PB-HSPC showing specific lineage scores detected at the end of the single-cell in vitro differentiation assay. GO-BP, gene ontology-biological processes; NES, normalized enrichment score.
To complement our single-cell phenotypic and transcriptional characterization with functional data, we measured the differentiation output of HD PB- or BM-HSPC subsets (n = 4 and 3, respectively), by exploiting an optimized in vitro multilineage single-cell assay (supplemental Figure 7C; supplemental Methods). With the exception of lymphoid progenitors and EPs, PB-HSPC subsets showed higher clonal differentiation efficiency than BM-HSPCs (Figure 3E; supplemental Figure 7D-E). The output of 397 HSPCs was scored as unilineage, bilineage, and multilineage based on the differentiation toward lymphoid, myeloid, erythroid, and megakaryocytic lineages. We observed that PB-HSPCs were mainly categorized as unierythroid, mixed, and lymphoid/erythroid clones, whereas BM-HSPCs displayed higher frequencies of only myeloid and only lymphoid clones (Figure 3F; supplemental Figure 7F), reflecting the transcriptional cluster distribution in the 2 sources (Figure 2D). Moreover, cHSPCs showed a higher frequency of erythroid/mecagaryocytic (Ery/MK) clones.
Finally, in vivo transplantation of BM-HSPCs and cHSPCs in immunodeficient mice indicated a similar homing potential of the 2 sources (supplemental Figure 8A-C). In line with their preactivated state, cHSPCs displayed higher early myeloid, erythroid, and B-cell reconstitution properties than BM-HSPCs, which showed increased LT engraftment potential. These data suggest that cHSPCs are capable of remaining and proliferating in the murine BM upon initial engraftment (supplemental text; supplemental Figure 8).
In summary, we found a strong consistency among phenotypic, transcriptional, and functional commitment of HSPC subsets isolated from BM and PB, with PB-HSPCs displaying a preactivated state for faster differentiation in case of demand.
cHSPCs display transcriptional downregulation of molecules controlling BM retention
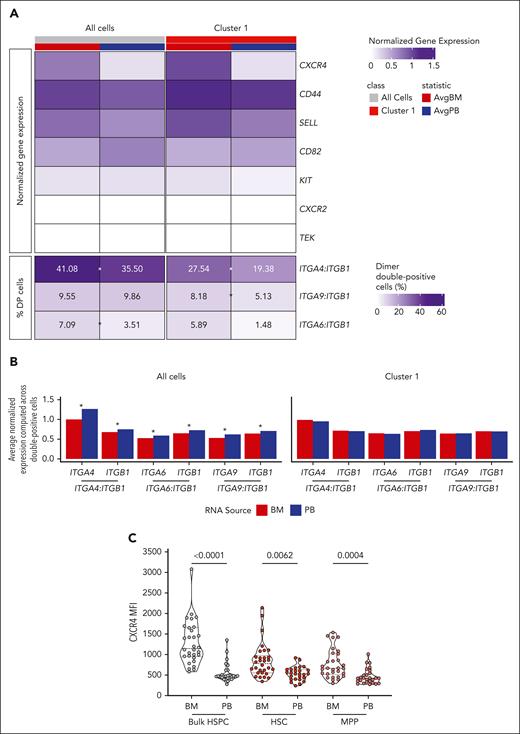
To investigate the molecular mechanisms driving human HSPC physiological egress from the BM, we evaluated the gene expression of 13 adhesion molecules involved in BM homing and retention, including CXCR4, CD82, and c-KIT, in total BM- or PB-HSPCs and in LT-HSCs (c1). Moreover, because VLA-4(ITGA4:ITGB1), α9β1(ITGA9:ITGB1), and α6β1(ITGA6:ITGB1) integrins are alpha/beta chain heterodimers, we assessed the percentage of cells coexpressing both chains.
We observed a higher expression of CXCR4, CD44, and SELL in BM-HSPCs than PB-HSPCs, whereas we measured no differences in c-KIT, CXCR2, and TEK expression between the 2 sources (Figure 4A). Moreover, resident-HSPCs showed a higher percentage of VLA4-, α9β1-, and α6β1-double positive (DP) cells with respect to cHSPCs. On the contrary, CD82 expression was enriched in PB-HSPCs, in line with its role in tuning cell migration41 (Figure 4A; supplemental Figure 8A). Remarkably, the expression of the single alpha and beta integrin chains within DP cells was similar in BM and PB LT-HSCs, whereas higher expression of the single chains was detected in DP total PB-HSPCs (Figure 4B). These transcriptional differences were also validated at protein levels, although reaching statistical significance only for CD44, CD82, ITGA6, and CXCR4, which showed the highest differential expression between the 2 HSPC sources (Figure 4C; supplemental Figure 9B).
cHSPCs display reduced expression of molecules controlling BM retention. (A) Heat map showing the average normalized gene expression of factors controlling BM retention (top) and the percentage of cells coexpressing genes encoding for integrins driving BM homing (dimer-DP cells) (bottom) in all cells as well as in cells from cluster 1 (LT-HSC) of BM (red) and PB (blue) origin. Asterisks show statistically significant differential frequencies of DP cells between the 2 sources (χ2 test, Bonferroni-adjusted P < .05). (B) Bar plots showing the average normalized expression of genes encoding for single integrin chains in ITGA4:ITGB1-, ITGA6:ITGB1-, and ITGA9:ITGB1-DP cells from total data set and LT-HSC (cluster 1) of BM (red) and PB (blue) origin. Asterisks show statistically significant differential gene expression between the 2 sources (2-sided Student t test, Bonferroni-adjusted P < .05). (C) Violin plots showing CXCR4 mean intensity of fluorescence (MFI) in total HSPCs, HSCs, and MPPs derived from HD-BM (n = 30) and -PB (n = 26). Mann-Whitney statistical test was applied for groups’ comparison and single P values are reported within the graph. AvgBM, average normalized gene expression in BM cells; AvgPB, average normalized gene expression in PB cells.
cHSPCs display reduced expression of molecules controlling BM retention. (A) Heat map showing the average normalized gene expression of factors controlling BM retention (top) and the percentage of cells coexpressing genes encoding for integrins driving BM homing (dimer-DP cells) (bottom) in all cells as well as in cells from cluster 1 (LT-HSC) of BM (red) and PB (blue) origin. Asterisks show statistically significant differential frequencies of DP cells between the 2 sources (χ2 test, Bonferroni-adjusted P < .05). (B) Bar plots showing the average normalized expression of genes encoding for single integrin chains in ITGA4:ITGB1-, ITGA6:ITGB1-, and ITGA9:ITGB1-DP cells from total data set and LT-HSC (cluster 1) of BM (red) and PB (blue) origin. Asterisks show statistically significant differential gene expression between the 2 sources (2-sided Student t test, Bonferroni-adjusted P < .05). (C) Violin plots showing CXCR4 mean intensity of fluorescence (MFI) in total HSPCs, HSCs, and MPPs derived from HD-BM (n = 30) and -PB (n = 26). Mann-Whitney statistical test was applied for groups’ comparison and single P values are reported within the graph. AvgBM, average normalized gene expression in BM cells; AvgPB, average normalized gene expression in PB cells.
cHSPC subsets display distinct migratory fate, with a priming of circulating lymphoid-HSPCs toward the thymus
To study the cHSPC migratory fate, we exploited published single-cell RNA sequencing (scRNAseq) datasets from different organs. Starting from 1 205 726 cells isolated from healthy human spleen, thymus, lung, liver, kidney, lymph nodes, and BM as control, we selected PTPRC and CD34–coexpressing cells as surrogate for human tissue-resident CD45+CD34+ cells, generating a human organ-resident (HuOR)–HSPC dataset (supplemental Table 13). For each HuOR, we defined an organ-specific module score based on the top 20 differentially expressed genes and assessed their expression in PB-HSPC subsets (Figure 5A). HuOR-BM score was expressed in primitive, myeloid, and lymphoid cHSPC subpopulations, indicating the BM origin of cHSPCs. On the contrary, extramedullary organ scores were detected in distinct PB-HSPC subsets, according to their migratory properties. Indeed, HuOR-spleen score was mainly expressed by transitioning MPPs/CMPs, whereas HuOR-thymus and HuOR–lymph node scores were mostly found within lymphoid subsets and MDPs. HuOR-LNG scores were mainly observed in MDPs and EPs, whereas HuOR-liver and HuOR-kidney scores were enriched in transitioning MPP/CMP, MDP, and Ery/MK subtypes (Figure 5A).
cHSPC subsets show differential migratory propensities toward extramedullary organs. (A) Heat map showing the scaled organ-specific module scores (top-20 DE genes, rows) evaluated for each PB-HSPC cluster (columns). (B) Heat map showing the scaled expression levels of TSP1 genes (rows) across MLPs (columns, clusters 4). Cells were clustered into K = 3 groups by K-means clustering algorithm. The table below displays the proportion of TSP1-high MLP in BM and PB. (C) Histograms showing the relative frequencies of the lymphoid cellular outputs of BM- and PB-MLPs detected at the end of the T-cell differentiation assay. The histogram above displays the proportions of CD4/CD8 double-negative (DN), CD4/CD8 double positive (DP), CD4 single-positive (CD4+ SP), and CD8 single-positive (CD8+ SP) cells detected within CD3+ cell compartment. The histogram below shows the proportions of T-cell precursors within the CD3– CD56– CD19– cell fraction. Data are shown as mean ± SEM. (D) UMAP embedding of the integrated scRNAseq data set, coloring cells by source: BM (red), PB (blue), and thymus (violet). (E) Density plots, showing cell classification as TSP1, TSP2, or ETP. (F) UMAP embedding, showing Monocle3 estimated pseudotime. (G) Percentages of TSP1 genes expressed (UMI > 0) by lymphoid–circulating CD34+ cells selected from a published peripheral blood mononuclear cells (PBMC) scRNAseq data set of healthy individuals from distinct ranges of age.47 The significance of the 1-way analysis of variance test across age classes revealed differences in the percentage of expressed TSP1 genes (P < .0001). These differences were then assessed through pairwise t test. Multiple hypotheses testing issue was accounted for adjusting the P values through the Benjamini-Hochberg method.
cHSPC subsets show differential migratory propensities toward extramedullary organs. (A) Heat map showing the scaled organ-specific module scores (top-20 DE genes, rows) evaluated for each PB-HSPC cluster (columns). (B) Heat map showing the scaled expression levels of TSP1 genes (rows) across MLPs (columns, clusters 4). Cells were clustered into K = 3 groups by K-means clustering algorithm. The table below displays the proportion of TSP1-high MLP in BM and PB. (C) Histograms showing the relative frequencies of the lymphoid cellular outputs of BM- and PB-MLPs detected at the end of the T-cell differentiation assay. The histogram above displays the proportions of CD4/CD8 double-negative (DN), CD4/CD8 double positive (DP), CD4 single-positive (CD4+ SP), and CD8 single-positive (CD8+ SP) cells detected within CD3+ cell compartment. The histogram below shows the proportions of T-cell precursors within the CD3– CD56– CD19– cell fraction. Data are shown as mean ± SEM. (D) UMAP embedding of the integrated scRNAseq data set, coloring cells by source: BM (red), PB (blue), and thymus (violet). (E) Density plots, showing cell classification as TSP1, TSP2, or ETP. (F) UMAP embedding, showing Monocle3 estimated pseudotime. (G) Percentages of TSP1 genes expressed (UMI > 0) by lymphoid–circulating CD34+ cells selected from a published peripheral blood mononuclear cells (PBMC) scRNAseq data set of healthy individuals from distinct ranges of age.47 The significance of the 1-way analysis of variance test across age classes revealed differences in the percentage of expressed TSP1 genes (P < .0001). These differences were then assessed through pairwise t test. Multiple hypotheses testing issue was accounted for adjusting the P values through the Benjamini-Hochberg method.
The enrichment of MLPs42-44 in cHSPCs and the higher expression of HuOR-thymus score in lymphoid-circulating subsets suggested the role of PB-MLPs in seeding the thymus. Indeed, we observed that a higher fraction of circulating MLPs (c4) displayed a gene signature of thymus seeding progenitor type 1 (TSP1; TSP1-high), described as a low-cycling immature population with thymus-emigrant properties45 (Figure 5B; supplemental Figure 10A), whereas few TSP1-high cells were found in BM-MLPs. However, TSP1-high MLPs from the 2 sources were transcriptionally consistent (supplemental Figure 10B) and showed an enrichment of ontologies related to cell migration compared with TSP1-low MLPs (supplemental Figure 9C). Moreover, TSP1 signature was found enriched also in PB compared with BM lymphoid progenitors in our multiome data set from HD-BM/PB-matched samples10 (supplemental Figure 10D). By comparing the differentially active domains of regulatory chromatine (DORC) of TSP1-high vs TSP1-low lymphoid progenitors, we found an enrichment of pathways associated with T-cell activation/differentiation and adhesion in TSP1-high cells, consistent with differential gene expression data (supplemental Figure 10C,E-F). In line with this finding, we measured a higher in vitro T-cell differentiation of PB-MLPs, which showed a increased production of terminally differentiated single-positive (SP) CD3+ T cells and committed CD7-high CD5+ T-cell progenitors with respect to BM-MLPs (Figure 5C).
Altogether, our results suggest that a fraction of MLPs in the BM acquires TSP1 signature and T-cell commitment to preferentially migrate into the PB to seed the thymus. To further investigate the role of PB-MLPs in connecting the BM and the thymus, we performed pseudotime analyses on an integrated scRNAseq data set encompassing BM-MLPs and PB-MLPs from our data set together with TSP1, TSP2, and early T-cell progenitor thymocytes from published works,45,46 which firstly described the early T-cell lymphoid differentiation in human thymus. Pseudotime analyses showed a clear differentiation trajectory starting from BM-MLPs through PB TPS1-high MLPs and TSP1-high thymocytes up to TSP2-high and early T-cell progenitor thymocytes (Figure 5D-F), with a progressive reinforcement of genes involved in T-cell differentiation/proliferation (CD7, IGLL1, TOP2A, MKI67, AURKB, NUSAP1) (supplemental Figure 10G). Finally, in accordance with the progressive reduction of thymic activity during aging,44 we found that TSP1 signature is reduced in circulating lymphoid progenitors from adult and older groups compared with the pediatric47 (Figure 5G).
Altogether, our data suggest that distinct cHSPC subsets are primed to seed diverse peripheral organs. Of note, trafficking T-cell–committed TSP1-high MLPs connect the BM with the thymus and actively contribute to T-cell lymphopoiesis.
Modeling in vivo trafficking in humans through IS analyses
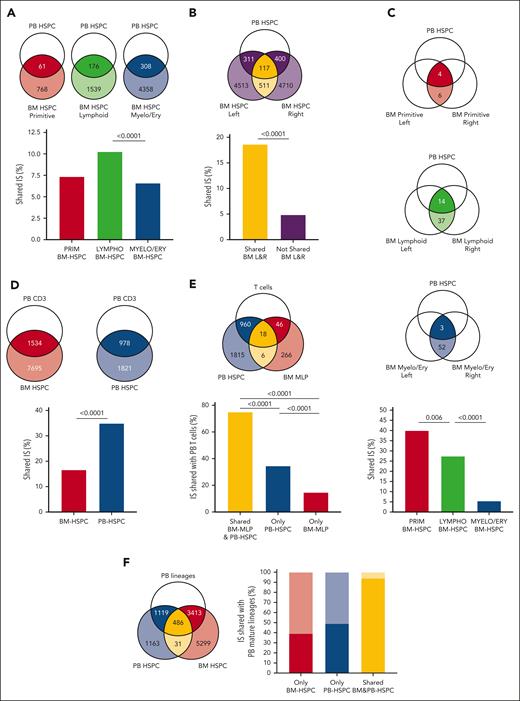
Vector IS analyses from patients with HSPC-GT are a powerful tool to investigate dynamics, hierarchical relationships, and hematopoietic output of HSPC subpopulations directly in vivo in humans.48-50 To study the in vivo trafficking of human HSPCs, we retrieved IS from BM-HSPCs collected from 2 distinct BM sites (left and right hips), PB-HSPCs, and mature PB lineages (CD14+, CD15+, CD3+, CD19+, CD56+ cells) isolated from 8 patients treated with HSPC-GT at steady-state hematopoiesis (>2 years after GT). For 4 of 8 patients, we collected ISs from PRIMITIVE (HSC + MPP), LYMPHOID (MLP + PreBNK), and MYELO/ERYTHROID (CMP, GMP, and MEP) BM-HSPC subsets from both BM sites (supplemental Table 14).
We first measured the propensity of BM-HSPC subsets to migrate into PB by means of shared IS with PB-HSPCs. We found that LYMPHOID BM-HSPCs showed a higher level of sharing with PB-HSPCs than PRIMITIVE and MYELO/ERYTHROID BM-HSPC subsets (Figure 6A), confirming the increased tendency of lymphoid progenitors to egress the BM (Figure 1I). Moreover, we detected ∼6.5% of clones shared between left and right BM-CD34+ cells (supplemental Figure 10A), which were also recaptured at higher frequencies in PB-HSPCs with respect to the not shared ones (Figure 6B), suggesting that cHSPCs recirculate among distant BM niches. The same analysis on BM-HSPC subsets revealed that IS shared between left and right PRIMITIVE BM-HSPCs showed the highest sharing with PB-HSPCs (Figure 6C), whereas IS not shared between distant niches displayed trends comparable with the overall population (Figure 6A; supplemental Figure 10B).
Modeling the in vivo trafficking of human HSPCs through IS clonal tracking. Venn diagrams show a schematic representation of the groups considered for the IS sharing analysis in each figure and report the IS number of the compared groups. (A) Histogram showing the relative frequencies of IS shared between BM PRIMITIVE (HSC + MPP, red), LYMPHOID (MLP + PreBNK, green), and MYELOID (CMP + GMP + MEP, blue) HSPC subsets and PB-HSPCs. (B) Histogram showing the relative frequencies of IS shared (yellow) and not shared (purple) between BM-HSPCs from 2 BM distant sites also recaptured in PB-HSPCs. (C) Histogram showing the relative frequencies of IS shared between BM PRIMITIVE (red), LYMPHOID (green), and MYELOID (blue) HSPC subsets from 2 BM distant sites also recaptured in PB-HSPCs. (D) Histogram showing the relative frequencies of IS shared between BM- or PB-HSPCs and PB T cells. (E) Histogram showing the relative frequencies of IS found in the shared fraction between BM-MLP and PB-HSPC (yellow), only in PB-HSPCs (blue) or only in BM-MLPs (red) that are also shared (dark colors) or not shared (light colors) with PB T cells. (F) Histogram showing the relative frequencies of IS found only in BM-HSPCs (red), only in PB-HSPCs (blue), and in the shared fraction between BM- and PB-HSPCs (yellow) that are also shared (dark colors) or not shared (light colors) with PB mature lineages. (G) Histograms showing the relative frequencies of IS found only in BM-HSPCs (red), only in PB-HSPCs (blue), and in the shared fraction between BM- and PB-HSPCs (yellow) that were detected in both myeloid and lymphoid (multilineage), in only myeloid (only myelo) and in only lymphoid (only lympho) PB mature cells. (H) Density plots representing the log10 sequence count distribution relative to IS found in BM-HSPCs (left) and in PB-HSPCs (right). The dot line refers to the clones shared between the 2 sources, whereas solid lines show the clonal abundance of not shared IS. Mann-Whitney test was performed to compare the sequence count distribution. For panels A-E,G, the P values shown in the figure were computed through Fisher exact test. Whenever >2 groups were compared, the nominal P values were adjusted with Bonferroni correction. Only significant P < .05 are shown in the plots.
Modeling the in vivo trafficking of human HSPCs through IS clonal tracking. Venn diagrams show a schematic representation of the groups considered for the IS sharing analysis in each figure and report the IS number of the compared groups. (A) Histogram showing the relative frequencies of IS shared between BM PRIMITIVE (HSC + MPP, red), LYMPHOID (MLP + PreBNK, green), and MYELOID (CMP + GMP + MEP, blue) HSPC subsets and PB-HSPCs. (B) Histogram showing the relative frequencies of IS shared (yellow) and not shared (purple) between BM-HSPCs from 2 BM distant sites also recaptured in PB-HSPCs. (C) Histogram showing the relative frequencies of IS shared between BM PRIMITIVE (red), LYMPHOID (green), and MYELOID (blue) HSPC subsets from 2 BM distant sites also recaptured in PB-HSPCs. (D) Histogram showing the relative frequencies of IS shared between BM- or PB-HSPCs and PB T cells. (E) Histogram showing the relative frequencies of IS found in the shared fraction between BM-MLP and PB-HSPC (yellow), only in PB-HSPCs (blue) or only in BM-MLPs (red) that are also shared (dark colors) or not shared (light colors) with PB T cells. (F) Histogram showing the relative frequencies of IS found only in BM-HSPCs (red), only in PB-HSPCs (blue), and in the shared fraction between BM- and PB-HSPCs (yellow) that are also shared (dark colors) or not shared (light colors) with PB mature lineages. (G) Histograms showing the relative frequencies of IS found only in BM-HSPCs (red), only in PB-HSPCs (blue), and in the shared fraction between BM- and PB-HSPCs (yellow) that were detected in both myeloid and lymphoid (multilineage), in only myeloid (only myelo) and in only lymphoid (only lympho) PB mature cells. (H) Density plots representing the log10 sequence count distribution relative to IS found in BM-HSPCs (left) and in PB-HSPCs (right). The dot line refers to the clones shared between the 2 sources, whereas solid lines show the clonal abundance of not shared IS. Mann-Whitney test was performed to compare the sequence count distribution. For panels A-E,G, the P values shown in the figure were computed through Fisher exact test. Whenever >2 groups were compared, the nominal P values were adjusted with Bonferroni correction. Only significant P < .05 are shown in the plots.
Given the enrichment of TSP1-high cells in cHSPCs, we assessed cHSPC in vivo contribution to T-cell lymphopoiesis by analyzing the IS sharing between trafficking- or resident-HSPCs and PB mature T cells. We observed that cHSPCs shared a higher number of ISs with T cells than their BM counterpart (Figure 6D). Moreover, BM-MLP clones that were also captured within PB-HSPCs display higher T-cell output with respect to resident BM-MLP clones, implying the direct link between BM-MLPs, cHSPCs, and T-cell production (Figure 6E). These data are in line with the hierarchical trajectory observed in our pseudotime analyses (Figure 5D-F).
Differently from the IS found only in BM-HSPCs or in PB-HSPCs, the clones shared between resident and circulating HSPCs were also highly detected in mature PB lineages (Figure 6F) and displayed multilineage profile (Figure 6G). Of note, BM-HSPC clones shared with cHSPCs displayed higher clonal abundance than clones found only in the resident-HSPC fraction. On the contrary, no differences were observed in the clonal size of IS belonging to cHSPCs that were shared or not with BM-HSPCs (Figure 6H). These findings imply that ISs shared between BM- and PB-HSPCs are capturing highly proliferating/differentiating clones with multilineage production in the BM.
Altogether, our IS data suggest that cHSPCs contribute to hematopoietic homeostasis and T-cell production, with HSC + MPP subsets showing the highest propensity to re-enter the BM, allowing for the connection of distant BM niches.
Discussion
Although the existence of cHSPCs has been long known,51-53 this population remains poorly characterized in humans, partly due to its rarity. Few works described the association of PB-CD34+ cell count with pathological conditions.3-9,54 Thanks to the advancement in single-cell technology, a recent study transcriptionally characterized human extramedullary HSPC in adult individuals, showing lineage priming and enrichment of an erythroid transcriptional signature in both splenic and circulating HSPCs.15 Nevertheless, cHSPC migratory fate and role in hematopoietic homeostasis in relationship with the BM-resident counterpart still remains unsolved. Here, we performed a comprehensive characterization of human cHSPCs, providing novel information over the previous literature.
By combining phenotypic, functional, and transcriptional single-cell analyses, we generated the largest reference dataset for studying human BM- and PB-HSPC subsets. Our data show a high concordance among phenotype, transcriptional commitment, and function of HSPC subsets isolated from 2 sources, highlighting their differential composition and biological role in hematopoietic homeostasis. In particular, the high proliferation and metabolic activity of BM-HSPCs imply their continuous support to hematopoietic turnover, whereas the preactivated state of PB-HSPCs suggests their role in patrolling peripheral tissues for on-demand rapid activation and local hematopoietic differentiation. Moreover, cHSPC low-replicative state could act as a protective mechanism toward multiple PB stimuli.
Several works reported the existence of HSPCs in peripheral organs in mice11-13 and humans,15-18 however, poor information exists on the mechanisms driving HSPC egress from the BM and the identity of cHSPC subpopulations with organ-specific seeding properties. The observed CXCR4 downregulation in PB-HSPCs (Figure 4) suggests a key role of this molecule in human HSPC recirculation. This finding is in line with previous studies in which HSPC circadian PB release was associated with stromal cell-derived factor 1 (SDF-1) fluctuations in the BM.55 Intriguingly, PB-HSPC oscillation displays consistent timing in humans and mice during the light-dark circle,56,57 implying similar clock of physiological recirculation in mammals. On the other hand, the upregulation of CD82 in PB-HSPC is in line with their enriched expression of macrocategories associated with cell adhesion and might explain the conserved homing potential of CXCR4-low PB-HSPCs (Figure 4; supplemental Figures 8B-C and 9B).
Our functional characterization and the HuOR data set generation were instrumental to gain insights into the destination of cHSPC subsets toward peripheral hematopoietic organs. In line with the published transcriptional similarity between PB and splenic HSC + MPP populations,15 PB-transitioning MPPs/CMPs showed the highest expression of HuOR-spleen module score, suggesting that trafficking multipotent progenitors are directed to the spleen to locally sustain extramedullary hematopoiesis (Figure 5). Additionally, the enrichment of phenotypic and transcriptional MLPs in PB-HSPCs (Figure 1), and the higher transcriptional similarities between PB lymphoid progenitors and HuOR-thymus HSPCs (Figure 5) suggest the role of these cells in seeding the thymus. This hypothesis is supported by the detection of a higher fraction of TSP1-high cells in circulating MLPs with transcriptional and functional T-cell commitment, as well as the progressive reinforcement of gene sets driving T-cell differentiation from BM-MLPs to thymic progenitors, through circulating MLPs. Finally, we measured, through ISs from patients with GT, a higher propensity of lymphoid progenitors to egress the BM (Figure 6A) and increased T-cell output of clones shared between cHSPCs and BM-resident MLPs (Figure 6E), providing definitive in vivo evidence on the role of circulating MLPs in T-cell lymphopoiesis.
Finally, HuOR data set unveiled unprecedented information on cHSPC destination toward nonhematopoietic tissues. Circulating myeloid progenitors might seed peripheral organs with high cell turnover, such as the lungs, for the replenishment of tissue-resident macrophages.58 Additionally, the similarity between trafficking MPP/CMP, myeloid and erythroid/MK progenitors, and resident-HSPCs from liver and kidney, 2 sites of embryonic hematopoiesis, might suggest both the existence of residual steady-state extramedullary hematopoiesis in these 2 organs in postnatal life and/or a residual exchange of progenitors among different hematopoietic niches to orchestrate a systemic response in case of stress conditions.59-61
IS clonal tracking has been exploited to investigate the dynamics and hierarchical relationship of human HSPCs in GT-treated patients.49,62,63 To our knowledge, this is the first time that IS are retrieved from distant BM niches in humans, allowing for the modeling of HSPC recirculation directly in vivo. The detection of shared HSPC clones between the 2 BM niches might be the result of (1) initial engraftment of 2 LT-HSCs in 2 distant niches, due to ex vivo proliferation of the same clone; or (2) a bystander trafficking of HSPC subpopulations in and out of the BM (recirculating clones). Our data supported this second hypothesis. Indeed, we found that IS shared between 2 BM distant sites were also recaptured at higher frequencies in PB than IS not shared between left and right BM niches (Figure 6B). Notably, HSCs/MPPs showed the highest sharing with these recirculating clones, implying their higher propensity to re-enter the BM with respect to other circulating progenitors (Figure 6C). This latter finding is also consistent with the observation that HuOR-BM score was the only HuOR module detected in circulating LT-HSCs (Figure 5A).
Our data also unveiled that ISs shared between BM and PB were marking multilineage clones actively contributing to hematopoiesis (Figure 6F-G). The higher clonal size of BM-HSPC clones recaptured in PB indicates that a fraction of cHSPCs derive from highly proliferating/differentiating BM-HSPC clones and might explain the preactivated state of cHSPCs. This finding also leaves open the possibility that the BM egress of these clones might be the result of a passive release along with mature differentiated cells, further highlighting the complexity of physiological HSPC trafficking.
Our results have also important translational implications. By analyzing >150 PB and BM samples from HDs of different ages, we unveiled high recirculation of HSC and MPP subsets in very young individuals (Figure 1) and the homing and multilineage properties of cHSPCs (supplemental Figure 8), suggesting that these cells might represent an alternative stem cell source for young infants. We have already shown that in vitro expanded cHSPCs may be exploited for GT applications for diseases with a high frequency of cHSPCs, such as in osteopetrosis.64 Future studies will be required to address the biological properties of pediatric cHSPCs and their potential clinical exploitation.
Additionally, the analyses of PB and BM samples from 35 patients affected by hematopoietic imbalances indicated that cHSPC composition is consistent with the associated hematopoietic disorder and with the composition of BM-HSPCs (Figure 1). In clinical practice, BM sampling is required for the diagnosis of hematopoietic diseases and the monitoring of the HSPC compartment upon treatments. Our data suggest the evaluation of cHSPCs as an easily accessible biomarker of BM-HSPCs, providing initial indications of the underlying disease, also to be exploited for research purposes, with limited access to BM samples.
In conclusion, our findings indicate that cHSPCs actively contribute to hematopoiesis by providing a steady-state reservoir of lineage-committed hematopoietic progenitors that can both recirculate among distant BM niches and migrate into different extramedullary organs to sustain hematopoietic homeostasis. Moreover, we propose cHSPCs as a biomarker of BM-HSPC state for translational studies, also providing insights on their homing and differentiation properties, highlighting the broad relevance of studying trafficking HSPCs.
Acknowledgments
The authors thank F. Ciceri and all the medical and nursing staff of the Pediatric Immunohematology and Bone Marrow Transplantation Unit of the San Raffaele Scientific Institute and the San Raffaele Stem Cell Programme; S. Zancan, M. Casiraghi, G. Tomaselli, and all San Raffaele Telethon Institute for Gene Therapy clinical trial office personnel for clinical trial management and support; the Orchard Therapeutics team for revising the manuscript; C. Waskow for her advice in handling NSGW41 mice; and C. Villa, E. Canonico, and S. Di Terlizzi of the Flow Cytometry Resource, Advanced Cytometry Technical Applications Laboratory at Ospedale San Raffaele for cell sorting and technical help with instrumentation. The authors are indebted to the patients and their families for their commitment.
This work was supported by Fondazione Telethon (TIGET Core Grant B2 to A.A. and TGT21016 to A.A. and S.S.), the Italian Ministero della Salute (grant GR-2019-12369499 to S.S.), the European Commission (grant ERARE-3-JTC 2015 EUROCID to A.A.) and the Else Kröner-Fresenius-Stiftung, Germany (prize to A.A.).
P.Q. conducted this study as partial fulfillment of her PhD in Molecular Medicine, Cellular and Molecular Biology Program, San Raffaele University, Milan, Italy. L.S. conducted this study as partial fulfillment of his PhD in Molecular Medicine, Gene and Cell Therapy program, San Raffaele University, Milan, Italy.
Authorship
Contribution: P.Q. performed phenotypic characterization, in vitro and in vivo assays, single-cell RNA sequencing (scRNAseq) library preparation, collected and interpreted the data, and wrote the manuscript; L.B.-R. performed phenotypic characterization and isolation of HSPC subpopulations, performed in vitro and in vivo assays, and analyzed the data; R.J.H. performed in vivo assay; M.M.N. contributed to scRNAseq library preparation and analyzed the data; G. Pacini and M.B. performed bioinformatic analyses on scRNAseq data; L.S. performed in vitro and in vivo experiments; G. Pais performed integration site (IS) analyses; F.B. performed IS retrieval through Sonication Linker-mediated-PCR (SLIM-PCR); C.P., A.R., and L.P. provided samples from neonates; F.D., I.M., and S.G. performed isolation of patient cell lineages; S.D., and F. Fraschetta contributed to patient management and support; G.B. provided healthy pediatric donor samples; M.O. and R.D.M. provided bone marrow (BM) and peripheral blood (PB) samples from older individuals; F.M. provided samples from neonates; E.M., and A.C. supervised IS analyses; M.E.B., F. Ferrua, V.C., and M.P.C. provided BM and PB samples from patients with Wiskott-Aldrich syndrome, adenosine deaminase deficiency, thalassemia, BM failure syndromes, metachromatic leukodystrophy, and mucopolysaccharidosis type 1-Hurler; B.G. provided BM and PB samples from patients with mucopolysaccharidosis type 1-Hurler and supervised scRNAseq analyses; I.Merelli supervised scRNAseq analyses; A.A. interpreted data, supervised the project, and revised the manuscript; and S.S. performed experiments, interpreted data, supervised the project, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Serena Scala, San Raffaele Telethon Institute for Gene Therapy, IRCCS San Raffaele Scientific Institute, via Olgettina 58, 20132 Milan, Italy; email: scala.serena@hsr.it.
References
Author notes
A.A. and S.S. contributed equally to this study.
All the data generated in this study have been deposited in the San Raffaele Open Research Data Repository https://data.mendeley.com/datasets/jywsy5zz96/1 Raw sequencing data are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE253485.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal