Background: In 2022, the WHO classified a new subtype of myelodysplastic neoplasms (MDS) with low blast count and SF3B1 mutation called MDS- SF3B1. SF3B1 is the major subunit of the SF3B spliceosome complex and is responsible for recognizing 3' splice sites in pre-mRNA for processing into mature mRNA. Mutations in SF3B1 have also been observed in other myeloid neoplasms. While MDS- SF3B1 has been associated with a better survival outcome, allelic variants such as SF3B1 K666N are associated with higher-risk MDS, transformation to AML and decreased OS. Additionally, SF3B1 E592K is more likely to be co-mutated with RUNX1, with a worse prognosis. Thus, not all SF3B1 allelic variants should be treated the same. The IPSS-M model incorporates SF3B1 mutations with different weights depending on co-mutations (i.e. isolated del(5q) or BCOR, BCORL1, RUNX1, NRAS, STAG2, SRSF2 mutations) but did not find a difference based on SF3B1 hotspot mutation. However, these and other studies have focused on specific amino acid substitutions but have not taken into consideration the protein domain structure of SF3B1. Using a large cohort of SF3B1-mutant myeloid malignancies, we determined whether HEAT repeat domain location was associated with differences between SF3B1-mutant myeloid neoplasms when comparing co-mutations and clinical characteristics.
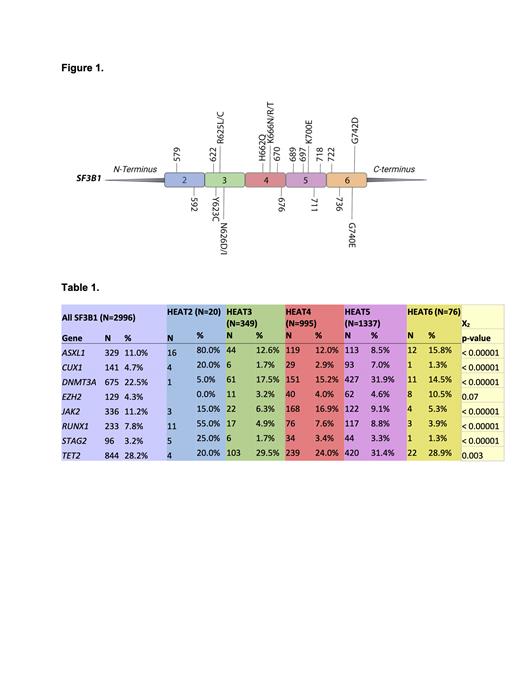
Methods: Bone marrow, peripheral blood, or FFPE tissue samples from a cohort of 2,996 unique patients with a suspected myeloid neoplasm from 6/30/2020-5/30/2023 were sequenced using a DNA 297 gene myeloid panel. We also analyzed a separate cohort of 112 patients from Sylvester Comprehensive Cancer Center (SCCC) and the University of Texas Southwestern Medical Center (UTSW) from 06/01/2017-06/30/2023 to validate the findings and assess for associations with clinical parameters such as blast count, time to disease progression, and survival. Statistics were performed using Chi-Square. SF3B1 has 22 HEAT repeats with SF3B1 mutations occurring predominately in HEAT Domains 2-6. For this study, we used UniProt definitions of amino acids 569-603 (HEAT2), 604-641 (HEAT3), 643-677 (HEAT4), 680-718 (HEAT5), and 763-801 (HEAT6) as shown in Figure 1.
Results: Out of 2,996 myeloid neoplasm patients with SF3B1 mutations, the SF3B1 allele with the highest prevalence was K700E, which falls into HEAT repeat number 5, followed by K666N/R/T and H662Q in HEAT repeat number 4, and R635C in HEAT repeat number 3. Mutations in HEAT repeat 2 and 6 were less common. Several genes were significantly co-mutated in different SF3B1 heat domains, including ASXL1 (11%, p-value =<0.00001), CUX1 (4.7%, p-value = <0.00001), DNMT3A (22.5%, p-value = <0.00001), EZH2 (4.3%, p-value = 0.07), JAK2 (11.2%, p-value= <0.00001), RUNX1 (7.8%, p-value = <0.00001), STAG2 (3.2%, p-value = <0.00001), and TET2 (28.2%, p-value = 0.003) shown in Table 1. The most common co-mutations within each HEAT domain were: HEAT2- ASXL1 (80.0%), RUNX1 (55%), and STAG2 (25%); HEAT3- TET2 (29.5%), DNMT3A (17.5%), and ASXL1 (12.6%); HEAT4- TET2 (24%), D NMT3A (15.2%), and JAK2 (16.9%); HEAT5- TET2 (31.4%) and DNMT3A (31%);and HEAT6- TET2 (28.9%), ASXL1 (15.8%), DMNT3A (14.5%), and EZH2(10.5%). In a separate cohort of 112 patients with clinical follow-up, 97 were diagnosed with MDS- SF3B1: 53 (55%) were male and the average age at diagnosis of 70 years (range: 27-89 years old). A total of 88 patients had 297 gene myeloid panel information, which revealed a similar HEAT repeat distribution: HEAT3 (19%), HEAT4 (27%), HEAT5 (45%), and HEAT6 (6%). The median OS of the entire cohort was 10.5 years (CI 95% 4.93-NE). With a median follow-up of 2 years; 75, 81, 63 and 80% of patients were alive in HEAT 3, 4, 5 and 6, respectively.
Conclusion: Distinct SF3B1 alleles defined by HEAT repeat location reflect distinct co-mutation patterns and may play a role in the biology of myeloid disorders. The relatively most enriched co-mutation patterns included: ASXL1 and RUNX1 in HEAT2, JAK2 in HEAT4, and EZH2 in HEAT6. Despite numerical differences in survival larger multicenter patient cohorts are needed to further define how distinct SF3B1 allelic variants and their HEAT repeat location correlate with prognosis and outcome in MDS and other myeloid neoplasms. In addition, further research is need into whether distinct SF3B1 alleles result in differences in RNA splicing signatures that may influence interactions with co-occurring mutations and disease biology.
Disclosures
Scarpa:Neogenomics Laboratories Inc.: Current Employment, Current holder of stock options in a privately-held company. Watts:Immune Systems Key: Research Funding; Takeda: Consultancy, Research Funding; Aptose: Consultancy; Rafael Pharma: Consultancy; Reven Pharma: Consultancy; Daiichi Sankyo: Consultancy; Servier: Consultancy; BMS: Consultancy; Rigel: Consultancy. Madanat:Taiho oncology: Honoraria; Stemline therapeutics: Honoraria; Morphosys: Honoraria, Other: travel reimbursement; Sierra Oncology: Honoraria; GERON: Consultancy; Novartis: Honoraria; OncLive: Honoraria; MD Education: Honoraria; Rigel Pharmaceuticals: Honoraria; Blueprint Medicines: Consultancy, Honoraria, Other: travel reimbursement. Sekeres:Kurome: Consultancy, Current holder of stock options in a privately-held company; Geron: Membership on an entity's Board of Directors or advisory committees; Novartis: Consultancy, Membership on an entity's Board of Directors or advisory committees; BMS: Consultancy, Membership on an entity's Board of Directors or advisory committees.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal