Introduction
Venetoclax (VEN) had been approved in Japan since March 2021 for acute myeloid leukemia (AML), and VEN-based regimens were widely used as initial therapy predominantly for elderly or unfit patients. Genetic mutations affect therapeutic outcomes of VEN-based therapy (CD DiNardo, et al. Blood. 2020). We performed gene panel sequencing using a next-generation sequencer for the newly diagnosed AML patients who were initially treated by VEN-based regimen and analyzed the relationship between gene mutations and outcomes in our regional cohort.
Patients and Methods
Hokkaido Leukemia Net (HLN) is prospective cohort study collecting acute leukemia samples from affiliated hospitals of the North Japan Hematology Study Group (NJHSG) covering Hokkaido and Iwate, Japan. We developed a compact AML panel covering mutation hot spots of 14 genes including TP53, CEBPA, NPM1, FLT3, KIT, NRAS, KRAS, CBL, PTPN11, DNMT3A, IDH1, IDH2, RUNX1 and ASXL1. Treatment efficacy was determined based on European LeukemiaNet 2017 response criteria in AML. This study was conducted in accordance with the Helsinki Declaration and was approved by the institutional review boards.
Results
A total of 89 AML cases registered in HLN from May 2021 to April 2023 were initially treated by VEN-based therapy (age range, 57-90 years; median age, 75 years; 49 males and 40 females, VEN+AZA 85, VEN+AraC 4). The median overall survival (OS) for all patients was 367 days (range 12-636).
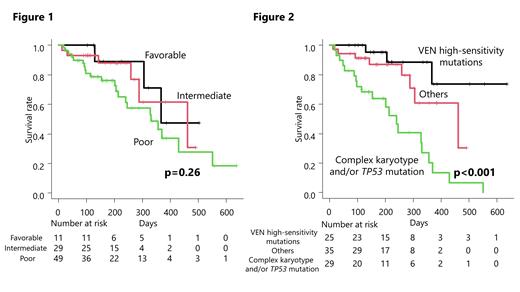
Patients who achieved complete remission (CR), CR with incomplete hematologic recovery (CRi), morphologic leukemia-free state (MLFS), or partial remission (PR) during the observation period and had no relapse were designated as Group 1, those who relapsed as Group 2, and those who became Stable disease (SD) or Progressive disease (PD) as Group 3. IDH2 mutations were significantly more frequent in Group 1 (P=0.010), and NPM1 mutations also tended to be more frequent in Group 1 and 2 (P=0.064). Furthermore, the ratio of CR or CRi was more than 80% in the patients with mutation of CEBPA-bZIP, DNMT3A, IDH1, IDH2, and NPM1 mutation-positive groups, respectively. These response groups clearly stratified prognosis. On the other hand, prognostic stratification according to biological disease factors depending on National Comprehensive Cancer Network (NCCN) 2017 stratification could not stratify the VEN-treated cohort (P=0.26) (Figure 1).
Next, the mutations with high CR+CRi rate ( CEBPA-bZIP, DNMT3A, IDH1, IDH2, and NPM1) were classified as “high-sensitivity mutations”. Patients with high-sensitivity mutations were significantly less likely to have complex karyotypes (P=0.026), but other patient characteristics such as age, sex, performance status, WBC count, other genetic mutations were not statistically different. Patients with high-sensitivity mutations had a significantly favorable OS than those without (median OS not reached; 95% CI, 367 - NA vs 331 days; 95% CI, 259-430, p=0.0022).
Univariate analysis was carried out for age, sex, karyotype, each gene mutation and “high-sensitivity mutations”. IDH2 mutation and high-sensitivity mutations were associated with favorable OS (P=0.016, 0.0022, log-rank test). As previously reported, complex karyotypes and TP53 mutations were associated with poor OS (p<0.001, <0.001, log-rank test).
Multivariate analysis including age, sex, “complex karyotype and/or TP53 mutation” and “high-sensitivity mutations” showed that the high-sensitivity mutations were independently associated with favorable OS (HR, 0.16; 95% CI, 0.053 to 0.62, p=0.0067; cox regression). Complex karyotype and/or TP53 mutation was independently associated with poor OS (HR, 2.39; 95% CI, 1.12 to 5.09, p=0.024; cox regression).
Based on these results, we were able to stratify the prognosis by dividing the patients into three groups according to the presence of high-sensitivity mutations, complex karyotype and/or TP53 mutations (Figure 2).
Conclusion
VEN-based therapy abrogates the efficacy of NCCN2017 stratification, however, the genetic mutation profile of the initial disease successfully stratified prognosis.
Disclosures
Kondo:Pfizer: Honoraria, Speakers Bureau; Novartis: Honoraria, Speakers Bureau; Otsuka Pharmaceuticals: Honoraria, Speakers Bureau. Iyama:Alexion Pharmaceuticals: Honoraria, Research Funding; MSD: Research Funding; Otsuka Pharmaceutical: Honoraria, Research Funding; Sanofi: Honoraria, Research Funding; SymBio Pharmaceuticals: Honoraria, Research Funding; Astellas: Honoraria; CSL Behring: Honoraria; Daiichi Sankyo: Honoraria; Meiji Pharma: Honoraria; Nippon Shinyaku: Honoraria; Novartis: Honoraria. Teshima:Meiji Seika Pharma: Membership on an entity's Board of Directors or advisory committees; DAIICHI SANKYO: Membership on an entity's Board of Directors or advisory committees; Sanofi: Membership on an entity's Board of Directors or advisory committees; Eisai: Research Funding; Sumitomo Pharma: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Janssen: Honoraria, Membership on an entity's Board of Directors or advisory committees; Novartis: Honoraria, Membership on an entity's Board of Directors or advisory committees; AbbVie: Honoraria; Roche Diagnostics: Membership on an entity's Board of Directors or advisory committees; Bristol-Myers Squibb: Honoraria; Merck Sharp & Dohme: Honoraria; Celgene: Honoraria, Membership on an entity's Board of Directors or advisory committees; AstraZeneca: Membership on an entity's Board of Directors or advisory committees; Takeda: Membership on an entity's Board of Directors or advisory committees; ONO: Research Funding; Asahi Kasei Pharma: Membership on an entity's Board of Directors or advisory committees, Research Funding; Fuji Pharma: Research Funding; NIPPON SHINYAKU: Honoraria, Research Funding; Otsuka: Research Funding; LUCA Science: Research Funding; Kyowa Kirin: Honoraria, Research Funding; Chugai: Honoraria, Research Funding; Priothera SAS: Research Funding; SHIONOGI: Research Funding; Astellas: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal