Abstract
This study reports the following: (1) frequency of TP53 comutation within each component of the European LeukemiaNet 2022 acute myeloid leukemia risk classification, (2) relevance of TP53 mutated variant allelic fraction <10%, (3) prognostic impact of −7, −5/del(5q), −17/abn(17p), complex karyotype/monosomal karyotype, or myelodysplasia-related gene mutations with/without mutated TP53.
TO THE EDITOR:
TP53 aberrations have been included as a specific diagnostic entity in the International Consensus Classification for acute myeloid leukemia (AML), emphasizing their importance as adverse risk markers.1 Although TP53 mutations are relatively common in therapy-related AML, most patients identified with TP53 variants have de novo disease, associated with chromosomal aneuploidies, including monosomy 7, monosomy 5/del(5q), or complex karyotype/monosomal karyotype (CK/MK).2 Several publications highlight poor outcomes for TP53-mutated AML, regardless of single- or multiple-hit allelic status.3-5 In 1 series, poor prognosis was associated with a TP53 allelic burden >40%.6 In the 2022 iteration of the European LeukemiaNet (ELN) risk recommendations for AML, adverse prognosis was classified by presence of poor risk gene fusions (listed in Table 1), monosomy 7, monosomy 5/del(5q), monosomy 17/abn(17p), CK/MK, or the newly described entity “AML with myelodysplasia-related (MR) gene mutations,” comprising ASXL1, BCOR, EZH2, RUNX1, SF3B1, SRSF2, STAG2, U2AF1, and/or ZRSR2 abnormalities. Presence of TP53 mutation with a variant allelic fraction (VAF) of at least 10% was also a requirement for ELN 2022 adverse risk. The VAF threshold of 10% was intended to counter the risk of ascribing adverse prognosis to a nonleukemic clonal hematopoietic variant.7
TP53 mutation frequency within ELN 2022 subgroups, according to de novo or secondary/therapy-related AML
| Classifier . | De novo AML (N = 2906) . | TP53 mutated, no. (%) . | TP53 wild type, no. (%) . | Secondary/therapy-related AML (n = 300) . | TP53 mutated, no. (%) . | TP53 wild type, no. (%) . |
|---|---|---|---|---|---|---|
| Favorable risk | 906 | 6 (0.7) | 900 (99.3) | 33 | 0 (0) | 33 (100) |
| RUNX1::RUNX1T1 | 164 | 2 (1.8) | 162 (98.2) | 6 | 0 (0) | 6 (100) |
| CBFB::MYH11 | 128 | 1 (0.8) | 127 (99.2) | 5 | 0 (0) | 5 (100) |
| Mutated NPM1 without FLT3-ITD | 415 | 1 (0.2) | 414 (99.8) | 21 | 0 (0) | 21 (100) |
| bZIP in-frame mutated CEBPA | 205 | 2 (1.0) | 203 (99) | 1 | 0 (0) | 1 (100) |
| Intermediate risk | 722 | 0 (0) | 722 (100) | 69 | 0 (0) | 69 (100) |
| Mutated NPM1 with FLT3-ITD | 257 | 0 (0) | 257 (100) | 8 | 0 (0) | 8 (100) |
| Wild-type NPM1 with FLT3-ITD (without adverse-risk genetic lesions) | 117 | 0 (0) | 117 (100) | 6 | 0 (0) | 6 (100) |
| MLLT3::KMT2A | 21 | 0 (0) | 21 (100) | 10 | 0 (0) | 10 (100) |
| Cytogenetic and/or molecular abnormalities not classified as favorable or adverse | 327 | 0 (0) | 327 (100) | 45 | 0 (0) | 45 (100) |
| Adverse risk | 1063 | 200 (18.8) | 863 (81.0) | 191 | 50 (26.2) | 141 (73.8) |
| DEK::NUP214 | 25 | 0 (0) | 25 (100) | 0 | 0 (0) | 0 (0) |
| KMT2A rearranged | 63 | 2 (3.2) | 60 (96.8) | 4 | 0 (0) | 4 (100) |
| BCR::ABL1 | 9 | 1 (11.1) | 8 (88.9) | 2 | 0 (0) | 2 (100) |
| KAT6A::CREBBP | 0 | 0 | 0 | 0 | 0 (0) | 0 (0) |
| GATA2, MECOM(EVI1) | 34 | 2 (5.9) | 32 (94.1) | 2 | 0 (0) | 2 (100) |
| MECOM(EVI1) rearranged | 10 | 1 (10.0) | 9 (90.0) | 9 | 4 (44.4)∗ | 5 (63.6) |
| Monosomy 5 or del(5q) | 198 | 133 (67.2) | 65 (32.8) | 51 | 29 (56.8) | 12 (23.5) |
| Monosomy 7 | 141 | 52 (36.9) | 89 (63.1) | 31 | 13 (41.9)∗ | 18 (58.1) |
| Monosomy 17/abn(17p) | 115 | 86 (70.4) | 34 (29.6) | 21 | 11 (52.4)∗ | 10 (47.6) |
| Complex and/or monosomal karyotype | 293 | 159 (54.2)∗ | 134 (45.7) | 67 | 39 (58.2)∗ | 28 (41.8) |
| AML with myelodysplasia-related gene mutations | 635 | 37 (5.8) | 598 (94.2) | 111 | 5 (4.5) | 106 (95.5) |
| ASXL1 | 214 | 9 (4.2) | 205 (95.8) | 46 | 1 (2.2) | 45 (97.8) |
| BCOR | 61 | 5 (8.2) | 56 (91.8) | 13 | 0 (0) | 13 (100) |
| EZH2 | 45 | 3 (6.7) | 42 (93.3) | 9 | 0 (0) | 9 (100) |
| RUNX1 | 284 | 8 (2.8) | 276 (97.2) | 40 | 1 (2.5) | 39 (97.5) |
| SF3B1 | 68 | 5 (7.4) | 63 (92.6) | 17 | 1 (5.9) | 16 (94.1) |
| SRSF2 | 103 | 0 (0) | 103 (100) | 29 | 1 (3.4) | 28 (96.6) |
| STAG2 | 106 | 3 (2.8) | 103 (97.2) | 22 | 0 (0) | 22 (100) |
| U2AF1 | 83 | 4 (4.8) | 79 (95.1) | 23 | 2 (8.7) | 21 (91.3) |
| ZRSR2 | 28 | 2 (7.1) | 26 (92.9) | 7 | 1 (14.3) | 6 (85.7) |
| Insufficient data to classify by ELN 2022 | 212 | 0 (0) | 212 (100) | 4 | 0 (0) | 4 (100) |
| Classifier . | De novo AML (N = 2906) . | TP53 mutated, no. (%) . | TP53 wild type, no. (%) . | Secondary/therapy-related AML (n = 300) . | TP53 mutated, no. (%) . | TP53 wild type, no. (%) . |
|---|---|---|---|---|---|---|
| Favorable risk | 906 | 6 (0.7) | 900 (99.3) | 33 | 0 (0) | 33 (100) |
| RUNX1::RUNX1T1 | 164 | 2 (1.8) | 162 (98.2) | 6 | 0 (0) | 6 (100) |
| CBFB::MYH11 | 128 | 1 (0.8) | 127 (99.2) | 5 | 0 (0) | 5 (100) |
| Mutated NPM1 without FLT3-ITD | 415 | 1 (0.2) | 414 (99.8) | 21 | 0 (0) | 21 (100) |
| bZIP in-frame mutated CEBPA | 205 | 2 (1.0) | 203 (99) | 1 | 0 (0) | 1 (100) |
| Intermediate risk | 722 | 0 (0) | 722 (100) | 69 | 0 (0) | 69 (100) |
| Mutated NPM1 with FLT3-ITD | 257 | 0 (0) | 257 (100) | 8 | 0 (0) | 8 (100) |
| Wild-type NPM1 with FLT3-ITD (without adverse-risk genetic lesions) | 117 | 0 (0) | 117 (100) | 6 | 0 (0) | 6 (100) |
| MLLT3::KMT2A | 21 | 0 (0) | 21 (100) | 10 | 0 (0) | 10 (100) |
| Cytogenetic and/or molecular abnormalities not classified as favorable or adverse | 327 | 0 (0) | 327 (100) | 45 | 0 (0) | 45 (100) |
| Adverse risk | 1063 | 200 (18.8) | 863 (81.0) | 191 | 50 (26.2) | 141 (73.8) |
| DEK::NUP214 | 25 | 0 (0) | 25 (100) | 0 | 0 (0) | 0 (0) |
| KMT2A rearranged | 63 | 2 (3.2) | 60 (96.8) | 4 | 0 (0) | 4 (100) |
| BCR::ABL1 | 9 | 1 (11.1) | 8 (88.9) | 2 | 0 (0) | 2 (100) |
| KAT6A::CREBBP | 0 | 0 | 0 | 0 | 0 (0) | 0 (0) |
| GATA2, MECOM(EVI1) | 34 | 2 (5.9) | 32 (94.1) | 2 | 0 (0) | 2 (100) |
| MECOM(EVI1) rearranged | 10 | 1 (10.0) | 9 (90.0) | 9 | 4 (44.4)∗ | 5 (63.6) |
| Monosomy 5 or del(5q) | 198 | 133 (67.2) | 65 (32.8) | 51 | 29 (56.8) | 12 (23.5) |
| Monosomy 7 | 141 | 52 (36.9) | 89 (63.1) | 31 | 13 (41.9)∗ | 18 (58.1) |
| Monosomy 17/abn(17p) | 115 | 86 (70.4) | 34 (29.6) | 21 | 11 (52.4)∗ | 10 (47.6) |
| Complex and/or monosomal karyotype | 293 | 159 (54.2)∗ | 134 (45.7) | 67 | 39 (58.2)∗ | 28 (41.8) |
| AML with myelodysplasia-related gene mutations | 635 | 37 (5.8) | 598 (94.2) | 111 | 5 (4.5) | 106 (95.5) |
| ASXL1 | 214 | 9 (4.2) | 205 (95.8) | 46 | 1 (2.2) | 45 (97.8) |
| BCOR | 61 | 5 (8.2) | 56 (91.8) | 13 | 0 (0) | 13 (100) |
| EZH2 | 45 | 3 (6.7) | 42 (93.3) | 9 | 0 (0) | 9 (100) |
| RUNX1 | 284 | 8 (2.8) | 276 (97.2) | 40 | 1 (2.5) | 39 (97.5) |
| SF3B1 | 68 | 5 (7.4) | 63 (92.6) | 17 | 1 (5.9) | 16 (94.1) |
| SRSF2 | 103 | 0 (0) | 103 (100) | 29 | 1 (3.4) | 28 (96.6) |
| STAG2 | 106 | 3 (2.8) | 103 (97.2) | 22 | 0 (0) | 22 (100) |
| U2AF1 | 83 | 4 (4.8) | 79 (95.1) | 23 | 2 (8.7) | 21 (91.3) |
| ZRSR2 | 28 | 2 (7.1) | 26 (92.9) | 7 | 1 (14.3) | 6 (85.7) |
| Insufficient data to classify by ELN 2022 | 212 | 0 (0) | 212 (100) | 4 | 0 (0) | 4 (100) |
bZIP, basic leucine zipper domain; ITD, internal tandem duplication.
The P value comparison between TP53 mutated and wild type was significant for all subgroups other than those asterisked, which had P > .05.
Previous reports indicated that adverse prognosis among patients with CK or monosomy 7 AML was limited to presence of comutated TP53.2,3,8 For most gene variants categorized as adverse prognosis in the ELN 2022 AML risk classification, however, the frequency and prognostic impact of cooccurring TP53 variants have not been determined. In addition, it remains to be confirmed whether AML with TP53 variant size <10% should be classified as nonadverse. Finally, the frequency and prognostic relevance of isolated monosomy 17/abn(17p) in the absence of a concurrent TP53 variant have not been comprehensively characterized.
We, therefore, sought to address the following queries in AML: (1) frequency of TP53 comutation with each component of the ELN 2022 risk classification, (2) prognostic relevance of TP53 abnormalities with VAF <10%, (3) prognostic relevance of monosomy 7, monosomy 5/del(5q), monosomy 17/abn(17p), or CK/MK in the absence of mutated TP53, and (4) prognostic relevance of AML with MR gene mutations without an overlapping TP53 variant.
Data were collated from the German Acute Myeloid Leukemia Study Group (European Genome-Phenome Archive; https://www.ebi.ac.uk/ega; under accession EGAS00001000275) and National Taiwan University Hospital (see supplemental Methods, available on the Blood website). Among the entire cohort of 3203 patients, 256 (8%) had a TP53 mutation. Although a TP53 mutation was more frequent in secondary vs de novo AML (16.8% vs 7.1%; P < .001), most patients with a TP53 mutation had de novo disease (80.4%) (Table 1). Further analysis was, therefore, focused on this subgroup.
Among patients with TP53-mutated de novo AML (206 patients), 86.1% received intensive chemotherapy, 7.6% received low-intensity therapy, and 6.3% received supportive care only. The median age of patients with a TP53 mutation was 60.5 years (range, 20-90 years), compared with 51.0 years (range, 1-98 years) for those who were TP53 wild type (P < .0001). An oncoprint characterizing the genomic landscape of TP53-mutated AML revealed a striking absence of mutations commonly recurrent in this disease (supplemental Figure 1). Lesions most frequently found in association with abnormal TP53 included CK (77%), monosomy 5/del(5q) (65%), monosomy 17/del(17p)/abn(17p) (40%), monosomy 7 (25%), and trisomy 8/del(8q) (22%). Almost complete overlap was noted between CK and MK (supplemental Figure 1).
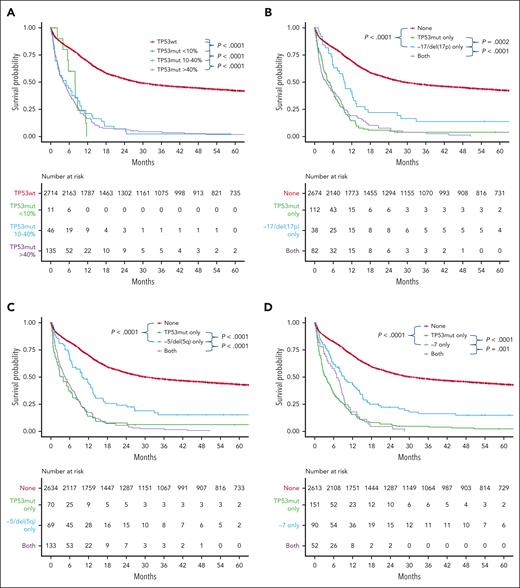
Patients with mutated TP53 had a median VAF of 58.1%. The proportions with VAF <10%, 10% to 40%, and >40% were 5.7%, 24.0%, and 70.3%, respectively. Although patients with VAF <10% appeared to have survival outcomes comparable to patients with higher TP53 VAF (median overall survival [OS], 7.89 vs 4.21 months; P = .6), a larger cohort will be required to confirm this finding (Figure 1A). In addition, our threshold for calling TP53 variants in this study was 5%. Therefore, the prognostic impact of TP53 variants below this threshold in AML remains to be explored. We next sought to determine the prognostic relevance of a TP53 mutation within each adverse risk subcategory where a cooccurring TP53 variant was present in at least 10 patients.
Survival outcome for TP53 mutated AML subgroups. Prognostic relevance of mutated TP53 according to (A) variant allele frequency and association with (B) monosomy 17/del(17p), (C) monosomy 5/del(5q), (D) monosomy 7, (E) CK/MK, or (F) an MR gene mutation among patients with de novo AML. Mut, mutated; wt, wild type.
Survival outcome for TP53 mutated AML subgroups. Prognostic relevance of mutated TP53 according to (A) variant allele frequency and association with (B) monosomy 17/del(17p), (C) monosomy 5/del(5q), (D) monosomy 7, (E) CK/MK, or (F) an MR gene mutation among patients with de novo AML. Mut, mutated; wt, wild type.
Among the subgroup with monosomy 17 and/or del(17p), TP53 was comutated in 66.7% (Table 1) and associated with poor prognosis (2-year OS, 7.2%; Figure 1B). Prognosis of monosomy 17 and/or del(17p) in the absence of a TP53 mutation was improved, but still poor (2-year OS, 22.0%). Among patients with monosomy 5 or del(5q), TP53 was concomitantly mutated in two-thirds of cases and conferred poor prognosis (2-year OS, 5.5%; Table 1). Although prognosis of monosomy 5 or del(5q) in the absence of mutated TP53 was better, OS remained poor (2-year OS, 24.1%; Figure 1C). A similar pattern was observed for patients with monosomy 7, in whom TP53 was frequently comutated (36.6%; Table 1) and associated with poor prognosis (2-year OS, 4.5%; Figure 1D). Similar to aneuploidies involving chromosome 17 and 5, prognosis for monosomy 7 in the absence of mutated TP53 was improved, but still poor (2-year OS, 22.3%).8 Consistent with prior reports, a TP53 mutation was common (52.8%) among patients with CK/MK and associated with poor prognosis (2-year OS, 4.6%). Compared with presence of a TP53 mutation, prognosis of CK/MK was improved in the absence of variant TP53, but still relatively poor (2-year OS, 30.4%) (Figure 1E). The new ELN 2022 adverse risk category “AML with MR gene mutations” was identified in 25.4% of the entire cohort (Table 1). Within this subgroup, TP53 was comutated in only 5.8% of cases and associated with poor prognosis (2-year OS, 6.2%; Figure 1F). In contrast, the prognosis of AML with MR gene mutations and wild-type TP53 was improved (2-year OS, 35.3%).
A notable finding was the poor prognosis conferred by a TP53 mutation, independent of associated aneuploidies or other adverse AML risk factors, including age >55 years, nonreceipt of intensive therapy or allogeneic hematopoietic cell transplant, or male sex (supplemental Tables 4 and 5). One limitation of this study was the small number of patients with secondary or therapy-related AML, preventing generalization of study findings to this population.
In conclusion, the presence of TP53 abnormalities confers adverse AML prognosis, regardless of the presence of other ELN 2022 adverse risk entities. TP53 variants are rare among patients outside ELN 2022 adverse risk. Although patients with TP53 VAF <10% occur uncommonly in AML, a larger cohort will be required to confirm poor prognosis in association with this subgroup. Although our findings support inclusion of monosomy 7, monosomy 5/del(5q), and monosomy 17/abn(17p) as adverse ELN 2022 classifiers in the absence of mutated TP53, our findings also support consideration of a TP53 mutation as a distinct AML entity, with “very-adverse” risk. Finally, for clinical trials exploring the role of novel drugs for patients with defective TP53, consideration should be given to also screening patients for isolated monosomy 17/17p, as a screening strategy limited to next-generation sequencing alone may exclude 16.4% of patients with only structural TP53 abnormalities.
Acknowledgments
This study was supported by fellowships and grants from the Australian National Health and Medical Research Council 2011139, 2006403, and 2018071; Medical Research Future Fund 2023403 and 1169950; Metcalf Family Foundation; Ministry of Science and Technology (Taiwan) (MOST 111-2314-B-002-279); and Ministry of Health and Welfare (Taiwan) (111-TDU-B-221-114001).
Authorship
Contribution: A.H.W. and R.M. designed the research; X.C.-H.T. collected the data; S.F., H.-A.H., and A.H.W. analyzed the data; and all authors wrote the manuscript and read, reviewed, and approved the manuscript.
Conflict-of-interest disclosure: A.H.W. has served on advisory boards for Novartis, AstraZeneca, Astellas, Janssen, Amgen, Roche, Pfizer, AbbVie, Servier, Gilead, BMS, Shoreline, Macrogenics, and Agios; receives research funding to the Institution from Novartis, AbbVie, Servier, Janssen, BMS, Syndax, Astex, AstraZeneca, and Amgen; serves on speaker's bureaus for AbbVie, Novartis, BMS, Servier, and Astellas, and is an employee of the Walter and Eliza Hall Institute (WEHI). WEHI receives milestone and royalty payments related to the development of venetoclax. Current and past employees of Walter and Eliza Hall Institute may be eligible for financial benefits related to these payments. The remaining authors declare no competing financial interests.
Correspondence: Andrew H. Wei, Peter MacCallum Cancer Centre and Royal Melbourne Hospital, 305 Grattan St, Melbourne, VIC 3000, Australia; e-mail: andrew.wei@petermac.org.
References
Author notes
∗S.F. and C.-H.T. are joint first authors.
†H.-A.H. and A.H.W. are joint senior authors.
Data are available on request from the corresponding author, Andrew H. Wei (wei.a@wehi.edu.au).
The online version of this article contains a data supplement.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal