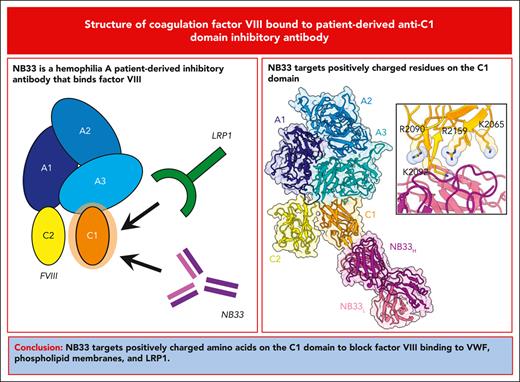
Key Points
The structure of FVIII bound to an anti–C1 domain antibody inhibitor reveals a novel epitope.
Antibody binding blocks multiple lysine and arginine residues implicated in FVIII endocytosis by dendritic cells.
Abstract
The development of pathogenic antibody inhibitors against coagulation factor VIII (FVIII) occurs in ∼30% of patients with congenital hemophilia A receiving FVIII replacement therapy, as well as in all cases of acquired hemophilia A. KM33 is an anti–C1 domain antibody inhibitor previously isolated from a patient with severe hemophilia A. In addition to potently blocking FVIII binding to von Willebrand factor and phospholipid surfaces, KM33 disrupts FVIII binding to lipoprotein receptor-related protein 1 (LRP1), which drives FVIII hepatic clearance and antigen presentation in dendritic cells. Here, we report on the structure of FVIII bound to NB33, a recombinant derivative of KM33, via single-particle cryo-electron microscopy. Structural analysis revealed that the NB33 epitope localizes to the FVIII residues R2090-S2094 and I2158-R2159, which constitute membrane-binding loops in the C1 domain. Further analysis revealed that multiple FVIII lysine and arginine residues, previously shown to mediate binding to LRP1, dock onto an acidic cleft at the NB33 variable domain interface, thus blocking a putative LRP1 binding site. Together, these results demonstrate a novel mechanism of FVIII inhibition by a patient-derived antibody inhibitor and provide structural evidence for engineering FVIII with reduced LRP1–mediated clearance.
Introduction
Hemophilia A is an X-linked recessive bleeding disorder afflicting 1 in 4065 male births worldwide and is characterized by defective or deficient coagulation factor VIII (FVIII), leading to uncontrolled bleeding events.1 The formation of pathogenic antibody inhibitors against FVIII occurs in 30% of patients with congenital hemophilia A receiving FVIII replacement therapy as well as in all cases of acquired hemophilia A.2 Most of the characterized antibody inhibitors target the A2, C1, and C2 domains of FVIII and disrupt coagulation mechanistically. Anti–C1 domain inhibitor antibodies can block FVIII binding to von Willebrand factor (VWF) and phospholipid surfaces, resulting in premature proteolytic degradation, clearance of FVIII, and/or impeding access to membrane surfaces, in which activated FVIII serves to nucleate the intrinsic tenase complex. KM33, a group AB anti–C1 domain antibody inhibitor was previously isolated from a patient with severe hemophilia A who presented with multiple inhibitor antibodies.3 In addition to potently inhibiting FVIII binding to VWF and phospholipid membranes, KM33 has the novel property of blocking the FVIII endocytosis by dendritic cells, which regulates the hepatic clearance of FVIII and antigen presentation.4,5 Studies incorporating site-directed mutagenesis with surface plasmon resonance (SPR) and live cell microscopy as well as hydrogen-deuterium exchange mass spectrometry (HDX-MS) have localized the KM33 epitope to several membrane-binding spikes in the C1 domain.5-11 To identify the amino acids that make up the KM33 epitope, this study reports on the structure of ET3i, a bioengineered human/porcine FVIII chimera, bound to a Fab fragment of NB33, a recombinant immunoglobulin G derivative of KM33, by single-particle cryo-electron microscopy (cryo-EM).
Study design
ET3i and NB33 were expressed, purified, and analyzed as previously described (see supplemental Methods, available on the Blood website).12-14 Cryo-EM sample preparation, data collection, image processing, and structure determination and validation are detailed in the supplemental Methods.
Results and discussion
Characterization of ET3i inhibition by NB33
Neutralization of ET3i by NB33 was assessed using a factor X activation chromogenic assay and Bethesda protocol. We determined an IC50 value of 3.64 nM in the chromogenic assay (supplemental Figure 1A) and a specific inhibitory activity of 7451 Bethesda units per mL NB33 in the Bethesda assay (supplemental Figure 1B), consistent the reports from previous studies.5,14 These results demonstrate that the inhibition of ET3i by NB33 is analogous to the inhibition of human FVIII by KM33.
Structure of ET3i:NB33 complex
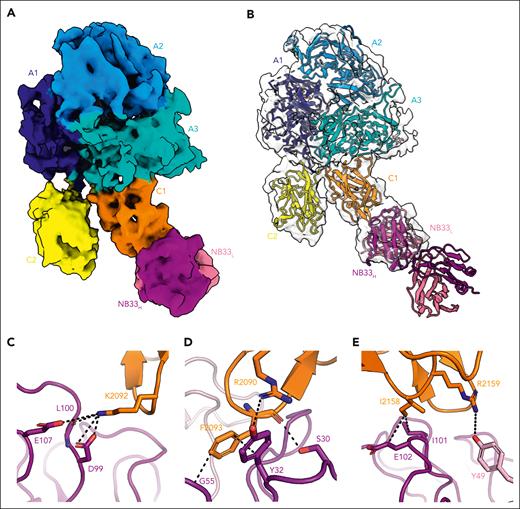
The structure of ET3i bound to NB33 was determined by single-particle cryo-EM at a nominal resolution of 4.23 Å (Figure 1A-B). The initial 2-dimensional classification showed intact particles with unambiguous densities in the NB33 Fab constant (heavy and light) and variable (heavy [VH] and light [VL]) domains (supplemental Figure 2A). The final sharpened map, excluding the flexible constant domains, displayed unequivocal densities for the A1-A2/A3-C1-C2 domains of ET3i and the variable domains of NB33. The ET3i A domains vary in local resolution, whereas the C2 domain has suboptimal resolution owing to its flexibility but sufficient density for complete model building. No large-scale conformational rearrangements of the C domains were observed as previously described.15,16
Cryo-EM structure of the ET3i:NB33 complex. Cryo-EM map (A) and structure (B) of ET3i bound to NB33 Fab fragment. (dark blue, porcine A1 domain; slate, human A2 domain; cyan, porcine A3 domain; orange, human C1 domain; yellow, human C2 domain; dark purple, NB33 heavy chain; and light pink, NB33 light chain). (C-E) Intermolecular contacts between ET3i residues (C-D) R2090-F2093 and (E) I2158-R2159 (orange) and NB33 heavy (dark purple) and light (light pink) chains. The dashed lines represent noncovalent interactions ≤5 Å.
Cryo-EM structure of the ET3i:NB33 complex. Cryo-EM map (A) and structure (B) of ET3i bound to NB33 Fab fragment. (dark blue, porcine A1 domain; slate, human A2 domain; cyan, porcine A3 domain; orange, human C1 domain; yellow, human C2 domain; dark purple, NB33 heavy chain; and light pink, NB33 light chain). (C-E) Intermolecular contacts between ET3i residues (C-D) R2090-F2093 and (E) I2158-R2159 (orange) and NB33 heavy (dark purple) and light (light pink) chains. The dashed lines represent noncovalent interactions ≤5 Å.
The ET3i:NB33 interface, which exhibits a local resolution of ∼3.5 to 4 Å (supplemental Figure 2C), is stabilized by a combination of electrostatic and hydrophobic interactions with a buried surface area of 822 Å2. The NB33 epitope centers on membrane-binding spikes in the C1 domain, composed FVIII residues R2090-S2094 and I2158-R2159.17 Residue K2092 anchors the C1 domain to NB33 by binding to an acidic cleft at the center of the paratope, forming multiple salt bridges and hydrogen bonds with the NB33 VH domain (Figure 1C). NB33 residue Y32 in the VH domain forms a hydrogen bond with FVIII residue R2090, in addition to hydrophobic interactions with residue F2093 (Figure 1D). These results are consistent with prior SPR and enzyme-linked immunosorbent assay data, demonstrating that the FVIII R2090A/K2092A/F2093A triple variant abrogated binding to KM33.8 The cryo-EM structure of BIVV001, a bioengineered extended half-life therapeutic FVIII fused to the VWF D′D3 domains, helped identify interactions between FVIII residues K2092 and F2093 and the D3 domain, supporting the obstruction of VWF binding as an inhibitory mechanism by KM33.5,18 Additional contacts were present between FVIII residues I2158-R2159 and the CDR3 loop of the NB33 VH domain (Figure 1E). Although previous HDX-MS data demonstrated interactions between KM33 and FVIII residues from 2077 to 2085, the ET3i:NB33 structure reveals no interactions with this region.5
Most of the intermolecular contacts are with the NB33 VH domain, consistent with the isolation of KM33 through genetic recombination from the VH gene segment of a patient with hemophilia A and combined with a noninhibitory immunoglobulin G4 VL.3 Recent pull-down assays illustrated that the KM33 VH domain, but not the VL domain, retained its affinity toward FVIII.19 However, several interactions are observed in the ET3i:NB33 cryo-EM structure at lower map contour levels between FVIII residues Y2043-Q2045 and the NB33 VL domain, as previously suggested by HDX-MS experiments.5 Together, the structure that we report is in agreement with previous studies demonstrating that the KM33 epitope overlaps with amino acids in the C1 domain, previously shown to bind VWF and/or phospholipid membranes.5,10,11 A complete list of intermolecular contacts is provided in supplemental Table 2.
NB33 and LRP1 bind to the FVIII lysine and arginine residues in the C1 domain
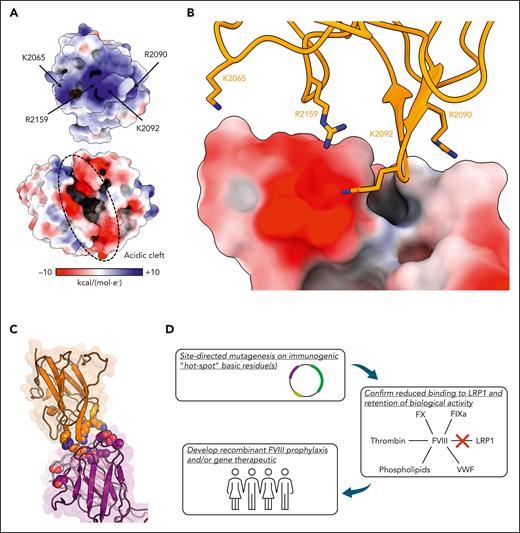
The ET3i:NB33 structure revealed a patch of positively charged residues in the FVIII C1 domain docked onto an acidic cleft formed by the VH/VL domain interface of NB33 (Figure 2A), providing the first structural insight into how KM33 blocks the uptake of FVIII by dendritic cells through low-density lipoprotein receptor-related protein 1 (LRP1)-mediated endocytosis. LRP1 uses clusters of extracellular complement-type repeat domains to bind and endocytose a wide variety of ligands, including FVIII, for processing and antigen presentation to T cells.20 Ligand binding to LRP1 occurs through a conserved acidic patch and an aromatic residue in the complement-type repeat domains, which target ligands carrying surface-exposed positively charged residues.21,22 The binding of LRP1 to FVIII is predicted to rely on an array of surface-exposed lysine residues on the FVIII light chain in a charge-dependent manner.10 Further studies have suggested a partial overlap between the LRP1 binding region and KM33 epitope.5,7,8,23 Our structural analysis of ET3i:NB33 was consistent with these experiments, revealing that residues K2065, R2090, K2092, and R2159 docked onto a patch of acidic NB33 residues, thus blocking the putative LRP1 binding region (Figure 2B-C). These positively charged FVIII residues have previously been shown to bind to VWF and/or phospholipid membranes, supporting the inhibitory mechanism of KM33 disruption of VWF and phospholipid interactions (supplemental Table 2).6,11,17,18 Intriguingly, NB33 appears to mimic the predicted LRP1 binding site using an aliphatic paratope to target positively charged FVIII residues and neighboring hydrophobic residues. These results provide structural evidence for the mechanism of FVIII clearance by LRP1 and suggest that mutating positively charged FVIII residues in the C1 domain can reduce hepatic clearance rates.
Electrostatic interactions between the C1 domain and NB33. (A) Electrostatic surface potential (blue, positive charge; red, negative charge) of the ET3i C1 domain (top) and NB33 Fab fragment (bottom) at ±10 kcal/(mol·e–). (B) FVIII residue K2065/R2090/K2092/R2159 (sticks) docked onto an acidic patch formed by the NB33 VH domain (surface). NB33 is colored based on the electrostatic surface potential, as in panel A. (C) The structure of the C1 domain bound to NB33. Charged residues in the epitope (C1 domain, orange) and paratope (NB33 VL, light pink; NB33 VH, dark purple) are depicted as spheres. (D) The proposed pipeline for the development of FVIII replacement therapeutics with reduced LRP1-mediated clearance and immunogenicity by targeting surface-exposed arginine and lysine residues.
Electrostatic interactions between the C1 domain and NB33. (A) Electrostatic surface potential (blue, positive charge; red, negative charge) of the ET3i C1 domain (top) and NB33 Fab fragment (bottom) at ±10 kcal/(mol·e–). (B) FVIII residue K2065/R2090/K2092/R2159 (sticks) docked onto an acidic patch formed by the NB33 VH domain (surface). NB33 is colored based on the electrostatic surface potential, as in panel A. (C) The structure of the C1 domain bound to NB33. Charged residues in the epitope (C1 domain, orange) and paratope (NB33 VL, light pink; NB33 VH, dark purple) are depicted as spheres. (D) The proposed pipeline for the development of FVIII replacement therapeutics with reduced LRP1-mediated clearance and immunogenicity by targeting surface-exposed arginine and lysine residues.
The structure of the C1 domain bound to NB33 bears resemblance to the crystal structure of the isolated C2 domain bound to BO2C11, a patient-derived anti–C2 domain antibody inhibitor that forms multiple salt bridges with FVIII residues R2215 and R2220 (supplemental Figure 4).24 Both antibodies target several β-hairpin loops and disrupt FVIII binding to VWF and phospholipid membranes, although the BO2C11 epitope was significantly more aliphatic than the electropositive KM33 epitope. In addition, the FVIII C1 domain forms extensive interactions with the VWF D′D3 domains in contrast to the C2 domain.18 Uptake of FVIII by monocyte-derived dendritic cells was blocked in the presence of BO2C11 or KM33 antibodies.25 Furthermore, the exposure of FVIII–/– mice to recombinant FVIII preincubated with BO2C11 or KM33 diminished the host immune response.7,25 Similar results were observed using a recombinant FVIII R2090A/K2092A/F2093A triple variant in the absence of antibodies, presumably because of the impaired binding to LRP1; however, the FVIII R2215A/R2220A double variant on the C2 domain showed no measurable effect on diminishing the immune response.8,25 Indeed, previous SPR studies have demonstrated that the C2 domain has no affinity for LRP1, suggesting that the BO2C11 antibody may indirectly block LRP1 binding through steric interference or overlap with the binding region for a non-LRP1 endocytic receptor.11 Although inhibitor development is a rare occurrence in cases of mild/moderate hemophilia A, our structural analysis may also indicate that certain missense mutations in the C1 domain may promote FVIII binding to LRP1 and induce an immune response.26 Together, these results suggest differential roles for positively charged residues in the C domains in driving LRP1-mediated FVIII endocytosis and immunogenicity.
In summary, the ET3i:NB33 structure represents the first structural analysis of a complex of a therapeutically active FVIII construct bound to a patient-derived pathogenic inhibitory antibody. Our structural analysis delineates the role of surface-exposed, positively charged residues on the C1 domain in binding to KM33, which disrupts the endocytosis of FVIII by dendritic cells. Fundamental questions concerning FVIII clearance require further investigation, including the role of non-LRP1 endocytic receptors, such as macrophage mannose receptors, which are unaffected by KM33-bound FVIII.7 These findings are critical for therapeutic strategies for designing a recombinant FVIII molecule with an extended half-life, reduced immunogenicity, and decreased clearance by dendritic cells (Figure 2D).
Acknowledgments
This work was supported by the National Hemophilia Foundation Judith Graham Pool Postdoctoral Research Fellowship (K.C.C.) and the National Institutes of Health (NIH), National Heart, Lung, and Blood Institute (award numbers R15HL135658 and U54HL141981 [P.C.S.] and award numbers R44HL117511, R44HL110448, U54HL112309, and U54HL141981 [C.B.D., P.L.]). A portion of this research was supported by the NIH National Institute of General Medical Sciences grant U24GM129547, performed at the Pacific Northwest Center for Cryo-EM at Oregon Health & Science University, and accessed through the Environmental Molecular Sciences Laboratory (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research.
Authorship
Contribution: K.C.C. planned and performed the experiments, analyzed the data, and assisted with writing the manuscript; N.G.A. performed the experiments and analyzed the data; K.A.E.A. assisted with data processing and interpretation; O.D. and R.M.H. performed the experiments and assisted with data processing and interpretation; C.B.D. and P.L. developed the expression and purification procedures for ET3i and assisted with writing the manuscript; C.H.C. developed the expression and purification procedures for NB33 and assisted with writing the manuscript; and P.C.S. planned the experiments, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: P.L. is listed as an inventor on a patent application describing ET3i and on patents owned by Emory University claiming compositions of matter that include modified FVIII proteins with reduced reactivity with anti-FVIII antibodies. C.B.D. and P.L. are cofounders of Expression Therapeutics and own equity in the company. Expression Therapeutics owns the intellectual property associated with ET3i. The terms of this arrangement have been reviewed and approved by Emory University in accordance with its conflict-of-interest policies. The remaining authors declare no competing financial interests.
Correspondence: P. Clint Spiegel Jr, Chemistry Department, Western Washington University, 516 High St, MS 9150 Bellingham, WA; e-mail: paul.spiegel@wwu.edu.
References
Author notes
Atomic coordinates for the factor VIII/NB33 structure are deposited in the Protein Data Bank (accession number 8G6I), and the cryo-electron microscopy maps are deposited in the Electron Microscopy Data Base (accession number EMD-29770).
Data are available on request from the corresponding author, P. Clint Spiegel Jr (paul.spiegel@wwu.edu).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal