Key Points
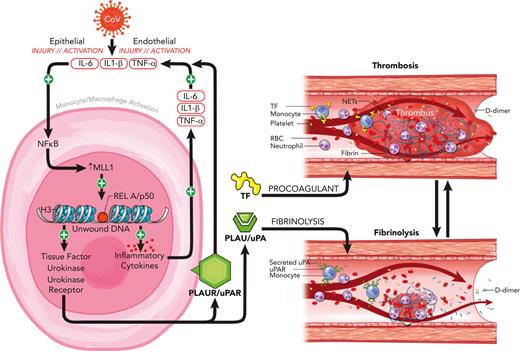
Monocyte/macrophage MLL1 promotes the expression of coagulopathy-related factors and proinflammatory cytokines after coronavirus infection.
Loss of monocyte/macrophage MLL1 attenuates the profibrinolytic and thrombophilic phenotype observed upon coronavirus infection in vivo.
Abstract
Coronavirus-associated coagulopathy (CAC) is a morbid and lethal sequela of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. CAC results from a perturbed balance between coagulation and fibrinolysis and occurs in conjunction with exaggerated activation of monocytes/macrophages (MO/Mφs), and the mechanisms that collectively govern this phenotype seen in CAC remain unclear. Here, using experimental models that use the murine betacoronavirus MHVA59, a well-established model of SARS-CoV-2 infection, we identify that the histone methyltransferase mixed lineage leukemia 1 (MLL1/KMT2A) is an important regulator of MO/Mφ expression of procoagulant and profibrinolytic factors such as tissue factor (F3; TF), urokinase (PLAU), and urokinase receptor (PLAUR) (herein, “coagulopathy-related factors”) in noninfected and infected cells. We show that MLL1 concurrently promotes the expression of the proinflammatory cytokines while suppressing the expression of interferon alfa (IFN-α), a well-known inducer of TF and PLAUR. Using in vitro models, we identify MLL1-dependent NF-κB/RelA–mediated transcription of these coagulation-related factors and identify a context-dependent, MLL1-independent role for RelA in the expression of these factors in vivo. As functional correlates for these findings, we demonstrate that the inflammatory, procoagulant, and profibrinolytic phenotypes seen in vivo after coronavirus infection were MLL1-dependent despite blunted Ifna induction in MO/Mφs. Finally, in an analysis of SARS-CoV-2 positive human samples, we identify differential upregulation of MLL1 and coagulopathy-related factor expression and activity in CD14+ MO/Mφs relative to noninfected and healthy controls. We also observed elevated plasma PLAU and TF activity in COVID-positive samples. Collectively, these findings highlight an important role for MO/Mφ MLL1 in promoting CAC and inflammation.
Introduction
Infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) results in physiologic derangements that stem from both the direct action of viral infection and ensuing host-immune response.1,2 Among these sequelae is coronavirus-associated coagulopathy (CAC), which results in increased thrombotic complications and mortality and features low-grade disseminated intravascular coagulopathy and thrombotic microangiopathy.3-5 The underlying pathophysiology is related in part to the combined actions of opposing processes, including thromboinflammation6,7 and altered fibrinolysis, because of the activity of urokinase (PLAU) and urokinase receptor (PLAUR), which results in D-dimer elevation that correlates with disease severity.3,8-11 The concurrent increased risk of arterial and venous micro/macrothrombosis represents a major unaddressed cause of SARS-CoV-2 morbidity and mortality.2,12
The pathophysiology of CAC involves exaggerated activation of leukocytes (including monocytes/macrophages [MO/Mφs]), endothelial cells, and platelets.2,13-16 MO/Mφs affect local and systemic coagulation and fibrinolysis via expression of tissue factor (F3; TF), PLAU, and PLAUR (“coagulopathy-related factors”) in response to various stimuli, including coronavirus infection and inflammatory cytokine and interferon/interferon receptor (IFN/IFNR) stimulation.17-24 The stimuli that govern MO/Mφ responsiveness to SARS-CoV-2 infection are not well understood but may be related to patient factors,25 alterations in IFN response,26,27 or epigenetic regulation by chromatin-modifying enzymes (CMEs) and associated proteins.28,29 CMEs alter MO/Mφ function and are implicated in a variety of disease contexts, including wound healing,30-32 atherosclerosis,33,34 and aneurysm development,35 and affect cytokine response in patients with diabetes after SARS-CoV-2 infection.29 Importantly, epigenetic changes may induce long-lasting alterations in gene expression (“epigenetic memory”) after acute infection and may facilitate the long-term sequelae seen after coronavirus infection.28,36
A candidate CME in altering MO/Mφ function is the histone methyltransferase mixed lineage leukemia 1 (MLL1/KMT2A), which is ubiquitously expressed in human tissues,37 functions in core complexes containing accessory proteins such as WDR538,39 and Menin,40 catalyzes the addition of methyl groups to lysine-4 residues on histone 3 proteins, and facilitates a chromatin conformation conducive for gene transcription.41 Initially recognized for its role in leukemogenesis,42 MLL1 has recently emerged as an important driver of the MO/Mφ response in many disease states.30,31,36,43,44 Previous work demonstrates MLL1-dependent interleukin 1β (IL-1β) expression and implicates epigenetic changes secondary to MLL1 suppression in MO/Mφs after recovery from sepsis that affect wound healing.36 MLL1 plays critical roles in facilitating immune responses downstream of proinflammatory and type I IFN signaling pathways involving IL-6, tumor necrosis factor α (TNFα), and STAT4.45,46 MLL1 is also important in orchestrating signaling involving NF-κB activation,47 which occurs through RelA activation and is induced by SARS-CoV-2 infection. Because RelA and IFN signaling regulate inflammatory cytokine and coagulopathy-related factor expression,24,26,48-50 we postulated that MLL1 may affect the expression of these factors in MO/Mφs to promote coagulopathy and systemic inflammation.
To this end, we used the murine betacoronavirus MHVA59 (a well-established model of SARS-CoV-2 infection29,51) to across in vitro and in vivo models to study the role of MLL1 in regulating the expression of CAC-related factors, inflammatory cytokines, and IFN/IFNRs. We found that infection of MO/Mφs yielded induction of MLL1, coagulopathy-related factors, and cytokines. Through loss-of-function models, we identified that the transcription of MLL1, these factors, and cytokines was directly regulated by MLL1. Next, we demonstrated that MLL1 is required for RelA-dependent transcription of coagulopathy-related factors in vitro. We showed that MLL1 is critical in inducing MO/Mφ and plasma expression of these factors and cytokines in response to coronavirus infection and in promoting a prothrombotic/profibrinolytic phenotype in vivo. Interestingly, loss of MO/Mφ MLL1 derepressed expression of Ifna, a mediator of coagulopathy in other contexts,26 despite attenuating coronavirus-induced coagulopathy. Finally, we observed upregulated MO/Mφ MLL1, coagulopathy-related factors, and inflammatory cytokine expression in CD14+ MO/Mφs and plasma derived from patients diagnosed as COVID-positive, who also displayed a concurrent induction of plasma PLAU and TF activity. Collectively, these results implicate MLL1 as a driver of MO/Mφ signaling, which is critical for CAC and inflammation.
Materials and methods
Animals and MHVA59 inoculation
C57BL6/J mice were obtained from The Jackson Laboratory and Kmt2afl/flLyz2Cre+/− and Kmt2afl/flLyz2Cre−/− mice were generated as previously described.36 MHVA59 was generated as described previously.29 Mice underwent intranasal inoculation with 2 × 105 plaque-forming units of MHVA59 or with phosphate-buffered saline. Animal studies were performed with the approval of the University of Michigan institutional animal care and use committee.
Tail bleeding assays and thromboelastography (TEG)
Mice were anesthetized using ketamine/xylazine and placed on a heating pad. Five millimeters of the tail tip was sharply excised, and the tail was immersed in saline at 37°C. Bleeding time was defined as cessation of bleeding for 1 minute. Rebleeds were identified if bleeding occurred within the 10-minute observation period for each animal. For TEG, whole blood was drawn from the inferior vena cava and citrate-anticoagulated (1:9; 3.2% sodium citrate: whole blood). Three hundred forty microliters of anticoagulated blood was mixed with 20 μL of 0.2 N CaCl2, and viscoelastic properties were analyzed using the Haemoscope TEG 5000 Thrombelastograph Hemostasis Analyzer (Haemonetics Corp). Where indicated, corn trypsin inhibitor (Prolytix) or TF-inhibiting antibody (TFI; rat-anti-mouse IgG2a/κ clone 1H1; Genentech) was incubated with anticoagulated whole blood for 15 minutes at 37°C before TEG. Plasma was obtained by centrifugation of whole blood at 2000g for 10 minutes for 2 sequential spins.
Human samples
Plasma/buffy coats were isolated from peripheral blood samples collected from hospitalized patients with and without COVID and from healthy controls by centrifugation of citrate-anticoagulated whole blood specimens for 2 sequential spins at 2000g for 15 minutes at room temperature. Patient characteristics are listed in supplemental Table 8, available on the Blood website. CD14+ MO/Mφs were isolated using the EasySep Human CD14 Positive Selection Kit (Stemcell Technologies). All samples were collected under approved protocols from the University of Michigan institutional review board.
Supplemental methods
A detailed description of other methods is provided in the supplement.
Results
Coronavirus infection of MO/Mφs induces MLL1, coagulopathy-related factors, inflammatory cytokines, and type I-III IFN/IFNRs
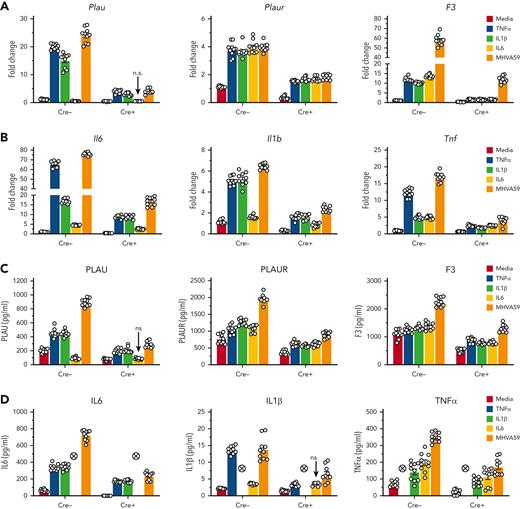
Previous studies demonstrate that inflammatory MO/Mφ activation occurs in an MLL1-dependent fashion.30,31,34,36 Therefore, we chose to investigate whether MLL1 expression increased in Mφs in response to coronavirus infection. We performed infection of bone marrow–derived Mφs (BMDMs) derived from C57BL6/J mice with murine coronavirus MHVA59. Infected BMDMs displayed induction of MLL1 expression relative to noninfected cells (Figure 1A-B). We observed similar upregulation of coagulopathy-related factors and inflammatory cytokine expression (Figure 1C-F) in BMDMs and in a second context that utilized immortalized murine Mφs (RAW264.7; supplemental Figure 1). In addition, we observed induction of messenger RNA (mRNA) levels of type I-III IFNs and IFNRs in BMDMs (supplemental Figure 2) in response to coronavirus infection. These results identify MLL1, the coagulopathy-related factors PLAU, PLAUR, and F3, the inflammatory cytokines IL-6, IL-1β, and TNFα, and type I-III IFNs as coronavirus-inducible factors in vitro.
Coronavirus infection of MO/Mφs induces expression of MLL1, coagulopathy-related factors, and inflammatory cytokines. BMDMs were harvested from C57Bl6/J mice (wild-type-BMDMs) and were infected with 1 MOI of the murine coronavirus MHVA59 for the indicated times, and mRNA (A) and protein levels (B; representative blot shown [β-actin served as loading control]) of Kmt2a/MLL1 were assayed by qRT-PCR and immunoblotting, respectively. (C) mRNA levels of factors important in CAC (Plau, Plaur, and F3) were measured in infected BMDMs. (D) mRNA levels of proinflammatory cytokines identified in the inflammatory signature resulting from acute SARS-CoV-2 (IL-6 and TNFα) and the MLL1-regulated cytokine IL-1β were measured in infected BMDMs. (E-F) Protein levels of coagulopathy-related factors (E) and proinflammatory cytokines (F) were assayed by ELISA. Bar graphs represent mean values from at least n = 5 independent experiments assayed in triplicate, and individual data points represent independent experiments. Errors bars represent SE. Statistical testing was performed using Kruskal-Wallis tests with corrections for multiple comparisons. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ELISA, enzyme-linked immunosorbent assay; MOI, multiplicity of infection; qRT, quantitative reverse transcription; SE, standard error.
Coronavirus infection of MO/Mφs induces expression of MLL1, coagulopathy-related factors, and inflammatory cytokines. BMDMs were harvested from C57Bl6/J mice (wild-type-BMDMs) and were infected with 1 MOI of the murine coronavirus MHVA59 for the indicated times, and mRNA (A) and protein levels (B; representative blot shown [β-actin served as loading control]) of Kmt2a/MLL1 were assayed by qRT-PCR and immunoblotting, respectively. (C) mRNA levels of factors important in CAC (Plau, Plaur, and F3) were measured in infected BMDMs. (D) mRNA levels of proinflammatory cytokines identified in the inflammatory signature resulting from acute SARS-CoV-2 (IL-6 and TNFα) and the MLL1-regulated cytokine IL-1β were measured in infected BMDMs. (E-F) Protein levels of coagulopathy-related factors (E) and proinflammatory cytokines (F) were assayed by ELISA. Bar graphs represent mean values from at least n = 5 independent experiments assayed in triplicate, and individual data points represent independent experiments. Errors bars represent SE. Statistical testing was performed using Kruskal-Wallis tests with corrections for multiple comparisons. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ELISA, enzyme-linked immunosorbent assay; MOI, multiplicity of infection; qRT, quantitative reverse transcription; SE, standard error.
MLL1 regulates basal expression of coagulopathy-related factors, inflammatory cytokines, and type I IFNs and type III IFNRs in MO/Mφs
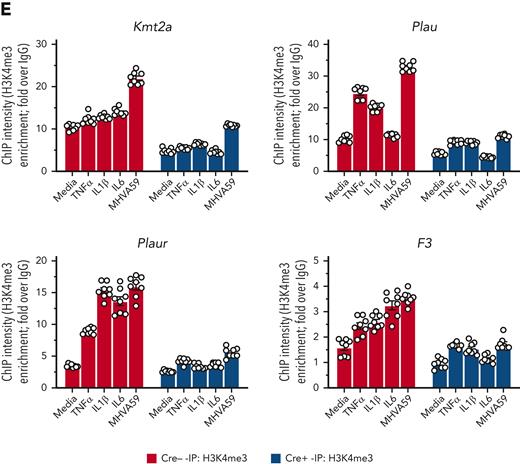
As MLL1 has been described to regulate inflammatory gene and type I IFN–dependent responses after toll-like receptor ligand and/or cytokine stimulation,31,45 we queried whether MLL1 was responsible for the expression of coagulopathy-related factors, inflammatory cytokines, and IFN/IFNR genes in MO/Mφs in the noninfected state. We analyzed expression of these genes in BMDMs from mice with myeloid-specific MLL1 knockout (Kmt2afl/flLyz2Cre+/−; denoted Cre+) and littermate controls (Kmt2afl/flLyz2Cre−/−; denoted Cre−). We confirmed the loss of MLL1 in harvested BMDMs (Figure 2A-B) and observed attenuated expression (Figure 2C,F) of coagulopathy-related factors and inflammatory cytokines in Cre+ BMDMs relative to Cre− cells. Furthermore, we observed MLL1-dependent H3K4me3 abundance on the promoters of coagulopathy-related factors (Figure 2G). Interestingly, denoting MLL1 self-regulation, MLL1 loss resulted in attenuated H3K4me3 abundance at its own promoter (Figure 2G). Moreover, baseline MLL1 promoter occupancy at candidate promoters in Cre− cells was observed, and as expected, this occupancy was lost in Cre+ cells (supplemental Figure 3A). Finally, we observed MLL1-dependent occupancy of phosphorylated RNA polymerase II β-subunit (Ser5; phospho-Rpb1), a marker of active transcription, at the promoters of coagulopathy-related factors (supplemental Figure 3B). These results indicate that MLL1 promotes the transcription of coagulopathy-related factors and itself.
MLL1 regulates the basal expression of coagulopathy-related factors and proinflammatory cytokines in BMDMs. BMDMs were harvested from mice carrying a myeloid-specific deletion of MLL1 (Kmt2afl/fl Lyz2Cre+/−; denoted Cre+) and littermate controls (Kmt2afl/fl Lyz2Cre−/−; denoted Cre−). (A) mRNA levels of Kmt2a were assayed in n = 8 Cre+ and Cre− animals analyzed in triplicate. (B) Protein levels of MLL1 were assayed in BMDMs from n = 4 Cre+ and Cre− mice by immunoblotting (representative blot shown [β-actin served as loading control]). (C-D) mRNA levels of coagulopathy-related factors (C) and proinflammatory cytokines (D) were assayed. (E-F) Protein levels of coagulopathy-related factors (E) and inflammatory cytokines (F) were measured by ELISA. (G) ChIP assays were performed using antibodies specific to either H3K4me3 or nontargeting species-specific IgG. Bars graphs represent mean values from at least n = 4 independent experiments with individual data points representing independent experiments. For ChIP experiments, bar graphs represent mean ChIP intensity relative to IgG derived from n = 8 samples performed in triplicate. Statistical analysis of pairwise comparisons was performed using Mann-Whitney tests. Error bars represent SE. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ChIP, chromatin immunoprecipitation; ELISA, enzyme-linked immunosorbent assay; IgG, immunoglobulin G; SE, standard error.
MLL1 regulates the basal expression of coagulopathy-related factors and proinflammatory cytokines in BMDMs. BMDMs were harvested from mice carrying a myeloid-specific deletion of MLL1 (Kmt2afl/fl Lyz2Cre+/−; denoted Cre+) and littermate controls (Kmt2afl/fl Lyz2Cre−/−; denoted Cre−). (A) mRNA levels of Kmt2a were assayed in n = 8 Cre+ and Cre− animals analyzed in triplicate. (B) Protein levels of MLL1 were assayed in BMDMs from n = 4 Cre+ and Cre− mice by immunoblotting (representative blot shown [β-actin served as loading control]). (C-D) mRNA levels of coagulopathy-related factors (C) and proinflammatory cytokines (D) were assayed. (E-F) Protein levels of coagulopathy-related factors (E) and inflammatory cytokines (F) were measured by ELISA. (G) ChIP assays were performed using antibodies specific to either H3K4me3 or nontargeting species-specific IgG. Bars graphs represent mean values from at least n = 4 independent experiments with individual data points representing independent experiments. For ChIP experiments, bar graphs represent mean ChIP intensity relative to IgG derived from n = 8 samples performed in triplicate. Statistical analysis of pairwise comparisons was performed using Mann-Whitney tests. Error bars represent SE. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ChIP, chromatin immunoprecipitation; ELISA, enzyme-linked immunosorbent assay; IgG, immunoglobulin G; SE, standard error.
We observed an induction of type I IFN (Ifna and Ifnb1) and type III IFNR levels in Cre+ cells compared with Cre− cells (supplemental Figure 4), and these results indicated a role for MLL1 in regulating basal IFN signaling and responsiveness in vitro. We observed similar MLL1-dependent regulation after siRNA-mediated MLL1 silencing in wild-type BMDMs (supplemental Figures 5-6) and RAW264.7 cells (supplemental Figure 7). Collectively, these results identify basal MLL1-dependent regulation of coagulopathy-related factors and inflammatory cytokines in MO/Mφs.
MLL1 regulates coronavirus and inflammatory cytokine-dependent induction of coagulopathy-related factors and inflammatory cytokines
In contrast to SARS-CoV-2, MHVA59 uses CEACAM152,53 (found on murine MO/Mφs) as a coreceptor for cell entry. Therefore, we sought to determine whether MLL1 mediated either MHVA59-initiated or MHVA59-independent induction of coagulopathy-related factors and inflammatory cytokines. We stimulated Cre− and Cre+ BMDMs with either cytokines induced in the post-coronavirus hyperimmune state54-56 or with MHVA59. We observed robust induction of coagulopathy-related factors and inflammatory cytokines after stimulation with each cytokine and MHVA59 infection in Cre− cells (Figure 3A-D). A notable exception was that the PLAU levels were suppressed by IL-6 stimulation, and this finding indicated the heterogeneity of cellular responses to inflammatory stimuli. We also observed MLL1-dependent enrichment of H3K4me3, MLL1, and phospho-Rpb1 on the promoters of MLL1 and coagulopathy-related factors after cytokine/coronavirus stimulation (Figure 3E; supplemental Figure 8A-B). Collectively, these results highlight an important role for MLL1 in cytokine/coronavirus-mediated induction of coagulopathy-related factors and inflammatory cytokines. We confirmed our findings in MLL1-silenced BMDMs (supplemental Figure 9) and RAW264.7 cells (supplemental Figure 10).
Loss of MLL1 attenuates coronavirus and proinflammatory cytokine-mediated induction of coagulopathy-related factors and proinflammatory cytokines in BMDMs. BMDMs were harvested from mice carrying a myeloid-specific deletion of MLL1 (Kmt2afl/fl Lyz2Cre+/−; denoted Cre+) and littermate controls (Kmt2afl/fl Lyz2Cre−/−; denoted Cre−). Cells were either stimulated with TNFα (5 ng/mL), IL-1β (5 ng/mL), or IL-6 (5 ng/mL) or infected with 1 MOI of MHVA59 for 24 hours. (A-B) mRNA levels of coagulopathy-associated factors and inflammatory cytokines were assayed in stimulated/infected cells by qRT-PCR. (C-D) Protein levels of coagulopathy-related factors (C) and inflammatory cytokines (D) were measured by ELISA. Crossed circles indicate cytokines, which was not measured in each assay as the particular cytokine was used for stimulation. (E) ChIP assays were performed using antibodies specific for H3K4me3 or with nontargeting species-specific IgG antibodies. ChIP intensity (relative to IgG) at the indicated promoters is shown. For qRT-PCR and ELISA experiments, graphs feature results from n = 10 animals assayed in triplicate. For ChIP experiments, bar graphs represent mean ChIP intensity relative to IgG from n = 8 animals assayed in triplicate. Error bars represent SE. For clarity of figure, graphical representation of statistical significance is not shown. Statistical analysis of data sets was performed using Mann-Whitney U tests and pairwise comparisons between Cre− and Cre+ cells were found to meet criteria for statistical significance (P < .05) except where indicated. ChIP, chromatin immunoprecipitation; ELISA, enzyme-linked immunosorbent assay; IgG, immunoglobulin G; MOI, multiplicity of infection; qRT, quantitative reverse transcription; SE, standard error.
Loss of MLL1 attenuates coronavirus and proinflammatory cytokine-mediated induction of coagulopathy-related factors and proinflammatory cytokines in BMDMs. BMDMs were harvested from mice carrying a myeloid-specific deletion of MLL1 (Kmt2afl/fl Lyz2Cre+/−; denoted Cre+) and littermate controls (Kmt2afl/fl Lyz2Cre−/−; denoted Cre−). Cells were either stimulated with TNFα (5 ng/mL), IL-1β (5 ng/mL), or IL-6 (5 ng/mL) or infected with 1 MOI of MHVA59 for 24 hours. (A-B) mRNA levels of coagulopathy-associated factors and inflammatory cytokines were assayed in stimulated/infected cells by qRT-PCR. (C-D) Protein levels of coagulopathy-related factors (C) and inflammatory cytokines (D) were measured by ELISA. Crossed circles indicate cytokines, which was not measured in each assay as the particular cytokine was used for stimulation. (E) ChIP assays were performed using antibodies specific for H3K4me3 or with nontargeting species-specific IgG antibodies. ChIP intensity (relative to IgG) at the indicated promoters is shown. For qRT-PCR and ELISA experiments, graphs feature results from n = 10 animals assayed in triplicate. For ChIP experiments, bar graphs represent mean ChIP intensity relative to IgG from n = 8 animals assayed in triplicate. Error bars represent SE. For clarity of figure, graphical representation of statistical significance is not shown. Statistical analysis of data sets was performed using Mann-Whitney U tests and pairwise comparisons between Cre− and Cre+ cells were found to meet criteria for statistical significance (P < .05) except where indicated. ChIP, chromatin immunoprecipitation; ELISA, enzyme-linked immunosorbent assay; IgG, immunoglobulin G; MOI, multiplicity of infection; qRT, quantitative reverse transcription; SE, standard error.
MLL1 alters IFN production and IFN-dependent coagulopathy-related factor expression in BMDMs
We chose to determine the role of MLL1 in mediating IFN production and responsiveness after coronavirus infection because type I IFNs such as IFN-α have been described to promote coagulopathy through the expression PLAUR24 and F3,57 and the dysregulation of IFN signaling has been implicated in the pathogenesis of SARS-CoV-2 sequelae.26,58,59 We observed that Cre+ BMDMs displayed an induction of Ifna, Ifnar1, Ifngr1, and type III IFN/IFNR mRNA expression but also a suppression of Ifng levels after coronavirus infection (supplemental Figure 11). Interestingly, though only IFN-α1 and IFN-γ stimulation induced Kmt2a expression (supplemental Figure 12A), only IFN-α1 stimulation yielded enhanced expression of coagulopathy-related factors, and this induction was blunted in Cre+ cells (supplemental Figure 12B-D). These results highlight a dominant role for MLL1 in regulating IFN-α/coagulopathy-related factor signaling after coronavirus infection in vitro.
MLL1 is required for RelA-dependent transcription of coronavirus-responsive factors
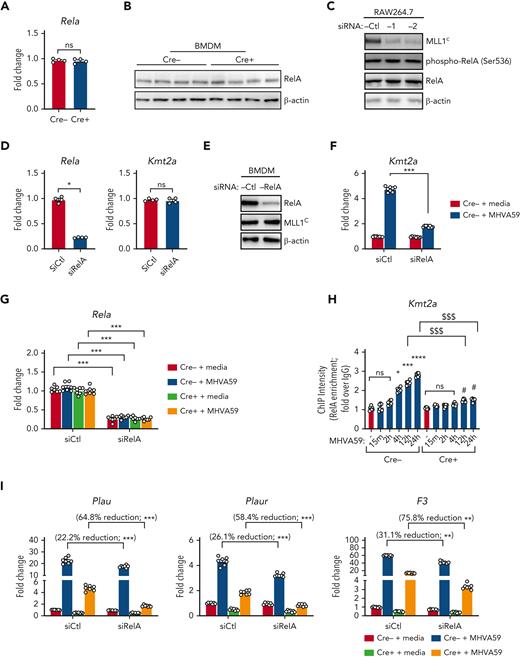
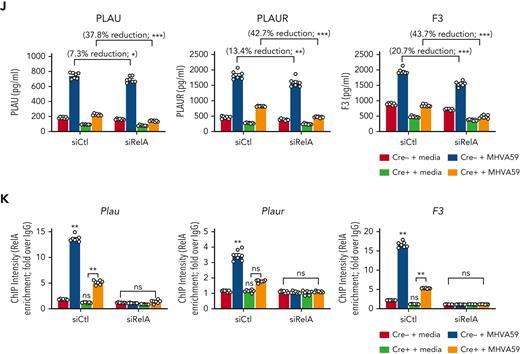
We chose to investigate collaborative gene regulation by MLL1 and RelA in MO/Mφs because MLL1 function has been previously shown to be influenced by NF-κB signaling.47 We did not observe altered levels of RelA mRNA or protein levels in Cre+ BMDMs relative to Cre− BMDMs (Figure 4A-B). Because activated RelA (phosphorylated on Ser276 or Ser536) protein levels were not readily detectable in BMDMs in the basal state, we analyzed RAW264.7 cells and did not observe any differences in phospho-RelA levels upon MLL1 knockdown (Figure 4C). Conversely, we did not observe RelA-mediated regulation of MLL1 expression in siRelA-treated BMDMs (Figure 4D-E). However, MHVA59 infection of RelA-silenced BMDMs yielded attenuated induction of MLL1 (Figure 4F), though RelA levels were not induced in either Cre− or Cre+ cells (Figure 4G). Collectively, these results identify that RelA is needed for maximal induction of MLL1 levels after coronavirus infection and that RelA and MLL1 do not participate in reciprocal regulation of each other’s expression in MO/Mφs in the basal state.
MLL1 is required for RelA-dependent transcription of coagulopathy-associated factors. (A) Rela mRNA levels were assayed in BMDMs harvested from MLL1 knockout mice (Cre+; n = 4) and littermate controls (Cre−; n = 4). (B) RelA protein levels are assayed in these cells by immunoblot (β-actin served as loading control). (C) The protein expression of MLL1 (MLL1C = C-terminal epitope), phospho-RelA (Ser536), and RelA was assayed in RAW264.7 cells that were transfected with either of 2 distinct siRNAs targeting MLL1 (denoted −1 and −2) or a nontargeting control (-Ctl). (β-actin served as loading control) (D) mRNA expression of Rela and Kmt2a was assayed in BMDMs transfected with siRNAs targeting Rela (smartpool; denoted siRelA) or a nontargeting control (siCtl). Results are representative of n = 4 independent experiments assayed in triplicate. (E) Protein levels of MLL1 and RelA were assayed, and representative immunoblot is shown. (F) BMDMs from littermate control mice (n = 8, assayed in triplicate) were transfected with the siRNAs and were infected with 1 MOI of MHVA59 for 24 hours, and mRNA levels of Kmt2a were measured. (G) Cre− and Cre+ BMDMs (n = 7 per group; assayed in triplicate) were transfected as indicated and infected with 1 MOI of MHVA59 for 24 hours and mRNA levels of Rela were assayed in these cells. (H) Cre− and Cre+ BMDMs (n = 7 per group; assayed in triplicate) were infected with MHVA59 for the indicated duration and ChIP assays were performed with antibodies specific to RelA or IgG. (I) mRNA levels of coagulopathy-associated factors were assayed in the indicated transfected cells (n = 7; performed in triplicate). (J) Protein levels of these coagulopathy-related factors were measured by ELISA (n = 7; performed in triplicate). (K) RelA ChIP assays were performed on candidate promoters and ChIP intensity relative to IgG was measured at the indicated promoters (n = 7 per group; assayed in triplicate). Bar graphs represent mean values and error bars represent SE. Statistical analysis was performed by either Mann-Whitney U test or Kruskal-Wallis test with corrections for multiple comparisons. Error bars represent SE. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ChIP, chromatin immunoprecipitation; ELISA, enzyme-linked immunosorbent assay; IgG, immunoglobulin G; MOI, multiplicity of infection; SE, standard error.
MLL1 is required for RelA-dependent transcription of coagulopathy-associated factors. (A) Rela mRNA levels were assayed in BMDMs harvested from MLL1 knockout mice (Cre+; n = 4) and littermate controls (Cre−; n = 4). (B) RelA protein levels are assayed in these cells by immunoblot (β-actin served as loading control). (C) The protein expression of MLL1 (MLL1C = C-terminal epitope), phospho-RelA (Ser536), and RelA was assayed in RAW264.7 cells that were transfected with either of 2 distinct siRNAs targeting MLL1 (denoted −1 and −2) or a nontargeting control (-Ctl). (β-actin served as loading control) (D) mRNA expression of Rela and Kmt2a was assayed in BMDMs transfected with siRNAs targeting Rela (smartpool; denoted siRelA) or a nontargeting control (siCtl). Results are representative of n = 4 independent experiments assayed in triplicate. (E) Protein levels of MLL1 and RelA were assayed, and representative immunoblot is shown. (F) BMDMs from littermate control mice (n = 8, assayed in triplicate) were transfected with the siRNAs and were infected with 1 MOI of MHVA59 for 24 hours, and mRNA levels of Kmt2a were measured. (G) Cre− and Cre+ BMDMs (n = 7 per group; assayed in triplicate) were transfected as indicated and infected with 1 MOI of MHVA59 for 24 hours and mRNA levels of Rela were assayed in these cells. (H) Cre− and Cre+ BMDMs (n = 7 per group; assayed in triplicate) were infected with MHVA59 for the indicated duration and ChIP assays were performed with antibodies specific to RelA or IgG. (I) mRNA levels of coagulopathy-associated factors were assayed in the indicated transfected cells (n = 7; performed in triplicate). (J) Protein levels of these coagulopathy-related factors were measured by ELISA (n = 7; performed in triplicate). (K) RelA ChIP assays were performed on candidate promoters and ChIP intensity relative to IgG was measured at the indicated promoters (n = 7 per group; assayed in triplicate). Bar graphs represent mean values and error bars represent SE. Statistical analysis was performed by either Mann-Whitney U test or Kruskal-Wallis test with corrections for multiple comparisons. Error bars represent SE. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ChIP, chromatin immunoprecipitation; ELISA, enzyme-linked immunosorbent assay; IgG, immunoglobulin G; MOI, multiplicity of infection; SE, standard error.
After coronavirus infection, we observed increased time-dependent RelA occupancy at the Kmt2a promoter (Figure 4H). The ability of RelA to occupy the MLL1 promoter was dependent on MLL1 expression, as attenuated promoter occupancy dynamics were observed in Cre+ cells. We also observed that, though RelA silencing alone yielded a modest suppression of the coagulopathy-related factors and inflammatory cytokine expression in response to MHVA59 infection (Figure 4I-J; supplemental Figure 13A-B), combined RelA and MLL1 loss resulted in robust abrogation of the coronavirus-dependent induction of these genes. Similarly, RelA abundance at the coagulopathy-related factor promoters was attenuated in Cre+ BMDMs that were infected with MHVA59 (Figure 4K). Collectively, these results show that MLL1 is required for RelA-dependent transcription of coagulopathy-related factors and inflammatory cytokines in MO/Mφs in vitro.
Coronavirus infection results in induction of MLL1 and dependent factors in vivo
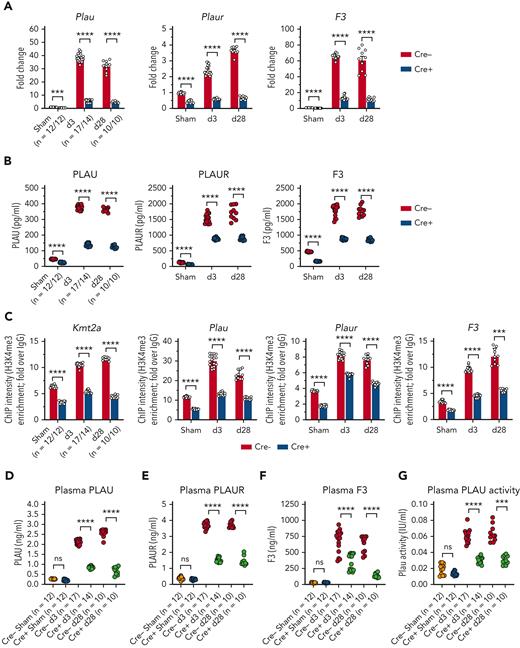
We next aimed to determine whether MLL1 and its dependent factors are induced in MO/Mφs upon coronavirus infection in vivo. C57BL6/J mice underwent intranasal inoculation with either 2 × 105 plaque-forming units of MHVA59 or with phosphate-buffered saline (sham). Through viral polymerase chain reaction (PCR) and MHVA59 enumeration assays, we determined that postinfection day 3 (d3) represented a period of robust viremia and infection of lung, splenic MO/Mφs, and BMDMs and that postinfection day 28 represented a late time point during which no measurable virus was present in these tissues (supplemental Figure 14). MHVA59 infection induced the expression of MLL1, coagulation-related factors, and inflammatory cytokines (Figure 5A-C; supplemental Figure 15A-B). Concordant with our in vitro findings, we observed induction of H3K4me3, MLL1, and RelA occupancy on the promoters of Kmt2a and the coagulopathy-related factors (Figure 5D; supplemental Figure 15C-D). Interestingly, MLL1 silencing in splenic MO/Mφs harvested from infected animals was able to attenuate coronavirus-mediated gene expression (supplemental Figure 16). However, compared with our in vitro studies (supplemental Figure 2), we observed that splenic MO/Mφs displayed suppressed levels of Ifnar1 and type II IFN/IFNRs, despite displaying inductions in Ifna and type III IFN/IFNRs (supplemental Figure 17). Nonetheless, we observed similar coagulopathy-related and inflammatory cytokine gene expression and epigenetic changes in BMDMs from infected animals (supplemental Figure 18).
Coronavirus induces the expression of MLL1 and its associated factors in MO/Mφs in vivo and promotes a prothrombotic and profibrinolytic phenotype. C57BL6/J mice underwent intranasal inoculation of 2 × 105 plaque-forming units (pfu) MHVA59 (postinfection day 3 mice [d3; n = 8]; postinfection day 28 mice [d28; n = 9]) or PBS (denoted as sham; n = 8). Mice were sacrificed at the indicated time points and plasma and splenic MO/Mφs (a surrogate for circulating MO/Mφs) were harvested. (A) Kmt2a mRNA levels were assayed by qRT-PCR. (B-C) The mRNA levels and protein levels of coagulopathy-associated factors in splenic MO/Mφs were measured by qRT-PCR and ELISA, respectively. (D) H3K4me3 abundance at the indicated promoters was assayed by ChIP assay. (E-F) Circulating levels of PLAU and PLAUR were measured by ELISA. (G) Plasma TF protein levels was measured by ELISA. (H) Plasma PLAU activity levels were measured using a colorimetric assay in which absorbance (A405) correlates with enzyme activity level through the cleavage of a plasmin (activated by PLAU) substrate that liberates p-nitroaniline. (I) Plasma TF activity was measured using a colorimetric assay in which the activation of factor X (FXa) by TF and factor VII (TF/FVIIa) and its cleavage of a FXa-specific substrate liberates p-nitroaniline. (J) The TF activity of lysed harvested splenic MO/Mφs was measured. (K) Tail vein bleeding time was measured in infected and sham mice. (L) The number of rebleeding events during tail vein bleeding time assays was tallied. (M) Whole blood was collected from infected and sham mice by inferior vena cava puncture and anticoagulated with 3.2% sodium citrate at a ratio of 9:1 (blood to citrate). TEG was performed and R time (time to formation of clot of 2 mm thickness) was measured (left panel). Representative TEGs are presented in the right panel. (N) To determine the role of TF in hypercoagulability as assayed by a shortened R time as measured by TEG after coronavirus infection, citrated whole blood samples from either sham or infected (d3) mice was treated with either corn trypsin inhibitor (CTI; 25 μg/mL final concentration) or a mouse specific anti-TF neutralizing antibody (TFI; clone 1H1 [Genentech]; 50 μg/mL final concentration) and the resultant viscoelastic properties were analyzed using TEG. Samples that were not subjected to treatment (NT) served as controls. Bar graphs represent mean values. qRT-PCR, ELISA, and ChIP data represent experiments performed in triplicate. Error bars represent SE. Statistical analysis of data sets was performed by either Mann-Whitney U test or Kruskal-Wallis test with corrections for multiple comparisons. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ChIP, chromatin immunoprecipitation; ELISA, enzyme-linked immunosorbent assay; PBS, phosphate-buffered saline; qRT, quantitative reverse transcription; SE, standard error
Coronavirus induces the expression of MLL1 and its associated factors in MO/Mφs in vivo and promotes a prothrombotic and profibrinolytic phenotype. C57BL6/J mice underwent intranasal inoculation of 2 × 105 plaque-forming units (pfu) MHVA59 (postinfection day 3 mice [d3; n = 8]; postinfection day 28 mice [d28; n = 9]) or PBS (denoted as sham; n = 8). Mice were sacrificed at the indicated time points and plasma and splenic MO/Mφs (a surrogate for circulating MO/Mφs) were harvested. (A) Kmt2a mRNA levels were assayed by qRT-PCR. (B-C) The mRNA levels and protein levels of coagulopathy-associated factors in splenic MO/Mφs were measured by qRT-PCR and ELISA, respectively. (D) H3K4me3 abundance at the indicated promoters was assayed by ChIP assay. (E-F) Circulating levels of PLAU and PLAUR were measured by ELISA. (G) Plasma TF protein levels was measured by ELISA. (H) Plasma PLAU activity levels were measured using a colorimetric assay in which absorbance (A405) correlates with enzyme activity level through the cleavage of a plasmin (activated by PLAU) substrate that liberates p-nitroaniline. (I) Plasma TF activity was measured using a colorimetric assay in which the activation of factor X (FXa) by TF and factor VII (TF/FVIIa) and its cleavage of a FXa-specific substrate liberates p-nitroaniline. (J) The TF activity of lysed harvested splenic MO/Mφs was measured. (K) Tail vein bleeding time was measured in infected and sham mice. (L) The number of rebleeding events during tail vein bleeding time assays was tallied. (M) Whole blood was collected from infected and sham mice by inferior vena cava puncture and anticoagulated with 3.2% sodium citrate at a ratio of 9:1 (blood to citrate). TEG was performed and R time (time to formation of clot of 2 mm thickness) was measured (left panel). Representative TEGs are presented in the right panel. (N) To determine the role of TF in hypercoagulability as assayed by a shortened R time as measured by TEG after coronavirus infection, citrated whole blood samples from either sham or infected (d3) mice was treated with either corn trypsin inhibitor (CTI; 25 μg/mL final concentration) or a mouse specific anti-TF neutralizing antibody (TFI; clone 1H1 [Genentech]; 50 μg/mL final concentration) and the resultant viscoelastic properties were analyzed using TEG. Samples that were not subjected to treatment (NT) served as controls. Bar graphs represent mean values. qRT-PCR, ELISA, and ChIP data represent experiments performed in triplicate. Error bars represent SE. Statistical analysis of data sets was performed by either Mann-Whitney U test or Kruskal-Wallis test with corrections for multiple comparisons. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ChIP, chromatin immunoprecipitation; ELISA, enzyme-linked immunosorbent assay; PBS, phosphate-buffered saline; qRT, quantitative reverse transcription; SE, standard error
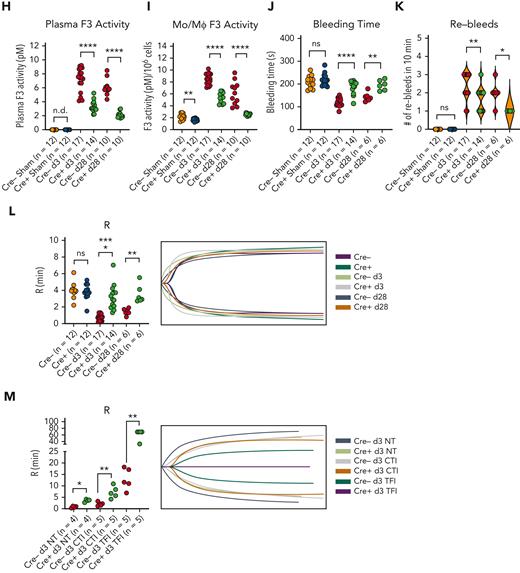
Finally, we observed coronavirus-mediated induction of plasma inflammatory cytokine and coagulopathy-related factor expression (supplemental Figure 15E; Figure 5) and soluble PLAU/PLAUR expression (Figure 5E-G). We also observed an induction in plasma PLAU activity and plasma and MO/Mφ TF activity, suggesting a concurrent fibrinolytic and hypercoagulable phenotype (Figure 5H-J). To explore the functional consequences of these results, we performed tail bleeding assays and TEG. Coronavirus infection resulted in faster cessation of tail bleeding and predisposed animals to a higher rate of rebleeding, thus demonstrating a hypercoagulable phenotype in which clot stability dynamics were perturbed, possibly through hyperfibrinolysis (Figure 5K-L). Using TEG, we observed that coronavirus infection yielded shortened clot formation times (R) (Figure 5M). We attributed this observation to TF activity as we observed a preferential effect of corn trypsin inhibitor treatment (an inhibitor of intrinsic coagulation)60 in prolonging R times in samples from sham animals (Figure 5N). Furthermore, treatment of samples with 100 μg/mL but not 50 μg/mL of murine-specific TF blocking antibody61 (TFI) was required to abrogate clot formation in infected samples, whereas 50 μg/mL of TFI was sufficient to block clot formation in sham samples (Figure 5N). Collectively, these results show that the expression of MLL1, coagulation-related factors, and inflammatory cytokines is induced in an MLL1-dependent manner in vivo in response to coronavirus infection and culminates in a prothrombotic and hyperfibrinolytic phenotype.
MLL1 loss in MO/Mφs attenuates coronavirus-mediated induction of coagulation-related factors and inflammatory cytokines in vivo
To further demonstrate MLL1-dependency for the induction of coagulation-related factors and inflammatory cytokines, we performed inoculation of Cre− and Cre+ mice with MHVA59. We observed robust induction of levels of coagulopathy-related factors and inflammatory cytokines in harvested Cre− BMDMs (supplemental Figure 19A-B) and splenic MO/Mφs compared with Cre+ cells (Figure 6A-B; supplemental Figure 20A-B). We also observed differentially induced H3K4me3 enrichment on the promoters of Kmt2a and the coagulopathy-related factors in Cre− splenic MO/Mφs and BMDMs relative to Cre+ cells upon coronavirus infection (Figure 6C; supplemental Figure 19C). However, we did not observe MLL1-dependence of RelA occupancy in splenic MO/Mφs or differential promoter occupancy of RelA in BMDMs (supplemental Figures 20C and 19D) in contrast to our results in vitro (Figure 4K).
Coronavirus-mediated induction of coagulopathy-related factors and the resultant coagulopathy is dependent on myeloid-specific expression of MLL1. Mice harboring a myeloid-specific MLL1 knockout (denoted Cre+; n indicated per group) and littermate controls (Cre−) underwent intranasal inoculation of 2 × 105 plaque-forming units (pfu) of MHVA59. Mice were sacrificed at the indicated time points and plasma and splenic MO/Mφs (a surrogate for circulating MO/Mφs) were harvested. (A-B) The mRNA and protein levels of coagulation associated factors were assayed by qRT-PCR and ELISA in splenic MO/Mφs, respectively. (C) H3K4me3 abundance at the indicated promoters was measured by ChIP assay. (D-E) Circulating levels of PLAU (D) and soluble PLAUR (E) were measured by ELISA. (F) Plasma PLAU activity levels were measured using a colorimetric assay in which absorbance (A405) correlates with enzyme activity level through the cleavage of a plasmin (activated by PLAU) substrate that liberates p-nitroaniline. (G) Plasma TF protein levels were measured by ELISA. (H) Plasma TF activity was measured using a colorimetric assay in which the activation of factor X (FXa) by TF and factor VII (TF/FVIIa) and its cleavage of a FXa-specific substrate liberates p-nitroaniline. (I) The TF activity of lysed harvested splenic MO/Mφs was measured. (J) Tail vein bleeding time was measured in infected and sham mice. (K) The number of rebleeding events during tail vein bleeding time assays was tallied. (L) Whole blood was collected from infected and sham mice by inferior vena cava puncture and anticoagulated with 3.2% sodium citrate at a ratio of 9:1 (blood to citrate). TEG was performed and R time (time to formation of clot of 2 mm thickness) was measured (left panel). Representative TEG is presented in the right panel. (M) To determine the role of TF in hypercoagulability as assayed by a shortened R time as measured by TEG after coronavirus infection, citrated whole blood samples from either infected (d3) Cre− or Cre+ mice was treated with either corn trypsin inhibitor (CTI; 25 μg/mL final concentration) or a mouse-specific anti-TF neutralizing antibody (TFI; clone 1H1 [Genentech]; 50 μg/mL final concentration) and the resultant viscoelastic properties were analyzed using TEG. Samples which were not subjected to treatment (NT) served as controls. Representative TEG is presented in the right panel. Bar graphs represent mean values and number of independent experiments per panel is as indicated. qRT-PCR, ELISA, and ChIP experiments were performed in triplicate. Error bars represent SE. Statistical comparisons were performed by either Mann-Whitney U test or Kruskal-Wallis test with corrections for multiple comparisons. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ChIP, chromatin immunoprecipitation; ELISA, enzyme-linked immunosorbent assay; qRT, quantitative reverse transcription; SE, standard error.
Coronavirus-mediated induction of coagulopathy-related factors and the resultant coagulopathy is dependent on myeloid-specific expression of MLL1. Mice harboring a myeloid-specific MLL1 knockout (denoted Cre+; n indicated per group) and littermate controls (Cre−) underwent intranasal inoculation of 2 × 105 plaque-forming units (pfu) of MHVA59. Mice were sacrificed at the indicated time points and plasma and splenic MO/Mφs (a surrogate for circulating MO/Mφs) were harvested. (A-B) The mRNA and protein levels of coagulation associated factors were assayed by qRT-PCR and ELISA in splenic MO/Mφs, respectively. (C) H3K4me3 abundance at the indicated promoters was measured by ChIP assay. (D-E) Circulating levels of PLAU (D) and soluble PLAUR (E) were measured by ELISA. (F) Plasma PLAU activity levels were measured using a colorimetric assay in which absorbance (A405) correlates with enzyme activity level through the cleavage of a plasmin (activated by PLAU) substrate that liberates p-nitroaniline. (G) Plasma TF protein levels were measured by ELISA. (H) Plasma TF activity was measured using a colorimetric assay in which the activation of factor X (FXa) by TF and factor VII (TF/FVIIa) and its cleavage of a FXa-specific substrate liberates p-nitroaniline. (I) The TF activity of lysed harvested splenic MO/Mφs was measured. (J) Tail vein bleeding time was measured in infected and sham mice. (K) The number of rebleeding events during tail vein bleeding time assays was tallied. (L) Whole blood was collected from infected and sham mice by inferior vena cava puncture and anticoagulated with 3.2% sodium citrate at a ratio of 9:1 (blood to citrate). TEG was performed and R time (time to formation of clot of 2 mm thickness) was measured (left panel). Representative TEG is presented in the right panel. (M) To determine the role of TF in hypercoagulability as assayed by a shortened R time as measured by TEG after coronavirus infection, citrated whole blood samples from either infected (d3) Cre− or Cre+ mice was treated with either corn trypsin inhibitor (CTI; 25 μg/mL final concentration) or a mouse-specific anti-TF neutralizing antibody (TFI; clone 1H1 [Genentech]; 50 μg/mL final concentration) and the resultant viscoelastic properties were analyzed using TEG. Samples which were not subjected to treatment (NT) served as controls. Representative TEG is presented in the right panel. Bar graphs represent mean values and number of independent experiments per panel is as indicated. qRT-PCR, ELISA, and ChIP experiments were performed in triplicate. Error bars represent SE. Statistical comparisons were performed by either Mann-Whitney U test or Kruskal-Wallis test with corrections for multiple comparisons. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ChIP, chromatin immunoprecipitation; ELISA, enzyme-linked immunosorbent assay; qRT, quantitative reverse transcription; SE, standard error.
Indicating MLL1-dependence, plasma levels of inflammatory cytokines (supplemental Figure 20D) and coagulopathy-related factors (Figure 6D-F) were differentially upregulated in infected Cre− mice. Furthermore, we observed a robust increase in plasma PLAU activity and plasma and MO/Mφ TF activity (Figure 6G-I) in Cre− mice compared with Cre+ mice. Finally, infected Cre− mice displayed shortened tail bleeding times with increased rebleeding events (Figure 6J-K) and shortened R times, which were TF-dependent (Figure 6L-M). Interestingly, uninfected Cre− and Cre+ mice did not display such differences, suggesting a context-specific role for MO/Mφ MLL1 in vivo. These results further support MO/Mφ MLL1’s role in the pathogenesis of coronavirus-dependent inflammatory coagulopathy.
MLL1 loss alters IFN responsiveness of MO/Mφs but does not affect MHVA59 infection in vivo
We next chose to investigate whether MO/Mφ MLL1 affected IFN production and responsiveness after coronavirus infection in vivo. Analysis of type I-III IFN/IFNR mRNA transcript expression showed a consistent pattern of IFN/IFNR transcript changes in splenic MO/Mφs between infected Cre− and C57BL6/J animals (supplemental Figures 21 and 17). Interestingly, only Ifna and Ifnlr1 mRNA levels were found to be concordantly induced in uninfected Cre+ BMDMs in vitro and Cre+ splenic MO/Mφs derived from sham animals in vivo (supplemental Figures 11 and 21). Analysis of infected BMDMs in vitro and splenic MO/Mφs derived from infected animals also revealed concordant induction of Ifna, Ifnar1, and Ifnlr1 in Cre+ cells compared with Cre− cells (supplemental Figures 11 and 21). Finally, a limited transcriptomic analysis by quantitative reverse transcription-PCR array of IFN-signaling relevant genes revealed heterogeneous coronavirus-mediated responses between uninfected and infected Cre− and Cre+ splenic MO/Mφs (sham: Cre+ vs Cre− MO/Mφs – decreased expression of 29 out of 66 [43.9%, 1.25-fold average decrease in expression] and increased expression of 37 out of 66 [56.1%, 3.11-fold average increase in expression] of analyzed transcripts; d3 Cre+ vs d3 Cre− MO/Mφs – decreased expression of 35 out of 66 [53.0%, 1.73-fold average decrease in expression] and increased expression of 31 out of 66 [47.0%, 2.07-fold average increase in expression] of analyzed transcripts), but nonetheless implicated Ifna transcripts as MLL1-repressible elements in both uninfected and infected states (supplemental Figure 21G). Despite these results in Cre− and Cre+ animals, we did not observe differences in MHVA59 infection of lung, splenic MO/Mφs, BMDMs, or whole blood through viral PCR or viral enumeration assay (supplemental Figure 22). Thus, our results highlight MLL1-IFN-α signaling that may preferentially affect coagulopathy after coronavirus infection.
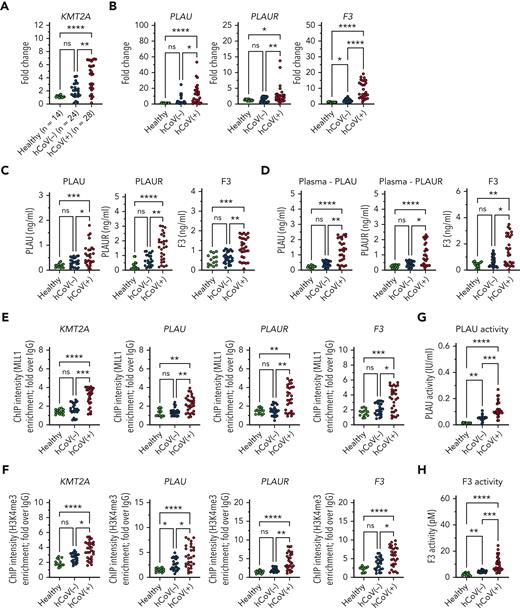
MLL1 and regulated factors are induced in CD14+ cells and plasma derived from patients infected with SARS-CoV-2 virus
We assessed whether MO/Mφs isolated from human SARS-CoV-2–positive samples displayed differential expression of MLL1 and dependent factors. We isolated CD14+ MO/Mφs and peripheral plasma samples from hospitalized patients with SARS-CoV-2 infection (n = 28; hCoV+), hospitalized patients without SARS-CoV-2 infection (n = 24; hCoV−), and healthy controls (n = 14) and identified elevated mRNA levels of MLL1 in hCoV+ patients compared with hCoV− patients and healthy controls (Figure 7A). We observed differential induction of MO/Mφ and plasma levels of coagulopathy-related factors in hCoV+ patients (Figure 7B-D; supplemental Figure 23A-C). We observed these changes in coagulopathy-related factors despite suppressed Ifna and Ifnb1 mRNA levels in CD14+ cells from hCoV+ patients compared with hCoV− patients (supplemental Figure 24). We also observed increased abundance of MLL1, H3K4Me3, and RelA occupancy at the promoters of MLL1 and coagulopathy-related factors in hCoV+ MO/Mφs relative to other groups (Figure 7E-F; supplemental Figure 23C-D). Finally, we observed that elevated plasma coagulation-related factor levels corresponded with induced PLAU and TF activity (Figure 7G-H). These data mirror our experimental findings regarding MLL1 and show that hCoV+ patients display a MO/Mφ and a plasma profile featuring induced coagulopathy-related factors and inflammatory cytokines.
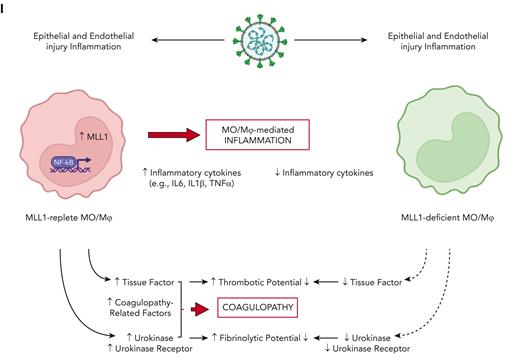
SARS-CoV-2–infected patients display elevated levels of MLL1 in MO/Mφs, and upregulated expression of MO/Mφ and circulating coagulopathy-associated factors and inflammatory cytokines. CD14+ cells were isolated peripheral blood samples from hCOV+ (n = 28), hCOV− (n = 24), and healthy controls (n = 14). (A) The mRNA levels of KMT2A were measured in isolated cells by qRT-PCR. (B-C) The mRNA and protein levels of coagulopathy-associated factors in MO/Mφs were assayed by qRT-PCR and ELISA, respectively. (D) The expression of coagulopathy-associated factors was measured from isolated plasma samples. (E) H3K4me3 and (F) MLL1 abundance at the indicated promoters was measured by ChIP assay. (G) Plasma PLAU activity was measured using a colorimetric assay in which absorbance (A405) correlates with enzyme activity level through the cleavage of a plasmin (activated by PLAU) substrate which liberates p-nitroaniline. (H) Plasma TF activity was measured using a colorimetric assay in which the activation of factor X (FXa) by TF and factor VII (TF/FVIIa) and its cleavage of a FXa-specific substrate liberates p-nitroaniline. (M) Schematic of the proposed mechanism of coronavirus-induced inflammation and coagulopathy as mediated by MLL1 in MO/Mφs. Bar graphs represent mean values. qRT-PCR, ELISA, and ChIP experiments were performed in triplicate. Error bars represent SE. Statistical comparisons were performed using the Kruskal-Wallis test with corrections for multiple comparisons. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ChIP, chromatin immunoprecipitation; ELISA, enzyme-linked immunosorbent assay; qRT, quantitative reverse transcription; SE, standard error.
SARS-CoV-2–infected patients display elevated levels of MLL1 in MO/Mφs, and upregulated expression of MO/Mφ and circulating coagulopathy-associated factors and inflammatory cytokines. CD14+ cells were isolated peripheral blood samples from hCOV+ (n = 28), hCOV− (n = 24), and healthy controls (n = 14). (A) The mRNA levels of KMT2A were measured in isolated cells by qRT-PCR. (B-C) The mRNA and protein levels of coagulopathy-associated factors in MO/Mφs were assayed by qRT-PCR and ELISA, respectively. (D) The expression of coagulopathy-associated factors was measured from isolated plasma samples. (E) H3K4me3 and (F) MLL1 abundance at the indicated promoters was measured by ChIP assay. (G) Plasma PLAU activity was measured using a colorimetric assay in which absorbance (A405) correlates with enzyme activity level through the cleavage of a plasmin (activated by PLAU) substrate which liberates p-nitroaniline. (H) Plasma TF activity was measured using a colorimetric assay in which the activation of factor X (FXa) by TF and factor VII (TF/FVIIa) and its cleavage of a FXa-specific substrate liberates p-nitroaniline. (M) Schematic of the proposed mechanism of coronavirus-induced inflammation and coagulopathy as mediated by MLL1 in MO/Mφs. Bar graphs represent mean values. qRT-PCR, ELISA, and ChIP experiments were performed in triplicate. Error bars represent SE. Statistical comparisons were performed using the Kruskal-Wallis test with corrections for multiple comparisons. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ChIP, chromatin immunoprecipitation; ELISA, enzyme-linked immunosorbent assay; qRT, quantitative reverse transcription; SE, standard error.
Discussion
Here, we report a critical role for MLL1 in promoting expression of coagulopathy-related factors and inflammatory cytokines after coronavirus infection and delineate a context-specific role for MLL1 in regulating RelA-dependent transcription of these factors (Figure 7I). We identify self-regulation of MLL1 expression and show that myeloid MLL1 loss attenuates the coronavirus-induced hypercoagulable/profibrinolytic phenotype in vivo despite derepressing the expression of IFN-α, which has been described as an inducer of coagulopathy after endotoxemia.57 Finally, we demonstrate differential expression/activity of MLL1 and coagulopathy-related factors in samples from patients with SARS-CoV-2 infection. These findings highlight a novel role for MO/Mφ MLL1 as a dominant regulator for the expression of factors important for CAC.
Because of the differences in cell specificity between MHVA59 (which uses CEACAM1 as coreceptors; found on murine MO/Mφs52,53) and SARS-CoV-2, distinguishing MHVA59-dependent regulation of MLL1 from virus-independent mechanisms is difficult. Nonetheless, we identify inflammatory cytokine–mediated MLL1 induction and reciprocal regulation of these cytokines by MLL1. Furthermore, we demonstrate Kmt2a and Plaur as 2 novel MLL1/RelA-regulated targets. Though we did not identify reciprocal regulation of the expression of MLL1 or RelA, we observed that MLL1 regulated RelA occupancy at the Kmt2a, Plau, Plaur, and F3 promoters in vitro but not in vivo. These results may highlight context-dependent NF-κB/RelA signaling dynamics, which have been described in response to a variety of stimuli.62-65 Nonetheless, we observed increased MLL1 and RELA occupancy on coagulopathy-related gene promoters in COVID-positive MO/Mφs relative to samples from patients without COVID patients and healthy controls, indicating a potential for collaborative transcriptional regulation after SARS-CoV-2 infection.
In contrast to the bidirectional regulation between MLL1 and its dependent inflammatory effectors, we observed the heterogeneity in MLL1-regulated IFN expression/responsiveness across experimental contexts, though Ifna suppression by MLL1 was a consistent observation. This finding may explain a mechanism by which type I IFN expression/responses are repressed after SARS-CoV-2 infection.26,27,58,59,66-68 Though type I IFNs or MLL1 may each promote the expression of coagulopathy-associated factors,24 our finding that MLL1 suppresses Ifna expression indicates an alternate mechanism for coagulopathy in a setting of diminished IFN signaling after coronavirus infection. Importantly, our observation of MLL1 as a mediator of IFN-α1–induced expression of coagulopathy-related factors in MO/Mφs may point to a role for MLL1 mediating IFN-dependent coagulopathy in other diseases such as sepsis. Nonetheless, additional studies are needed to delineate the relationship between MLL1 and global MO/Mφ transcription, as attenuated expression of inflammatory cytokines/coagulopathy-related factors after MLL1 loss may relate to a general decrease in transcription, with MLL1-Ifna regulation representing an exception to such a relationship.
Furthermore, an important role for CME and associated proteins continues to be described in the immunopathogenesis of SARS-CoV2 infection.28,29,69-72 We observed rapid and sustained upregulation in MLL1 expression/function after coronavirus infection. Our finding that coronavirus-induced gene expression in MO/Mφs is abrogated by silencing MLL1 is interesting as it suggests that MLL1 activity may enforce “epigenetic memory” after coronavirus infection; however, what contribution MLL1-associated proteins/MLL1-complex components play in acute and chronic responses remains unclear. As such memory is thought to contribute to the long-term sequelae in conditions such as aging, malignancy, and after recovery from sepsis, it is possible a prolonged state of smoldering inflammation/dysregulated coagulation, and other symptoms associated with “long-COVID”69-71 may be mediated through “epigenetic memory.”
The ability of CME and associated proteins to affect acute and chronic disease states renders factors such as MLL1 and its core components (eg, Menin, WDR5) as attractive therapeutic targets to blunt the sequelae of SARS-CoV-2 infection.38,40,42,73-77 Targeting MLL1/MLL1 complexes with small molecule inhibitors has been described for MLL1-rearranged leukemia treatment,40,74 and such inhibitors may be used to target MLL1-driven CAC, during which the balance between opposing procoagulant and fibrinolytic systems is perturbed. By addressing both hypercoagulability and hyperfibrinolysis, such therapies could prevent immunothrombosis/dysregulated fibrinolysis and restore a normal coagulation profile without the attendant risks of current anticoagulant therapies, especially in the intensive care unit patient population where the bleeding risk of anticoagulants has been shown to outweigh the benefits.78 Nonetheless, because MLL1 expression is not limited to MO/Mφs,37 it is unclear what effects would result from global inhibition of MLL1 activity (though immunosuppressive effects and poor wound healing a possible side-effects), and therefore MO/Mφs-targeted therapies must be designed. Furthermore, our experimental model of myeloid-specific MLL1 loss may capture MLL1’s effects on other cells of myeloid lineage, such as neutrophils, which have been implicated in SARS-CoV-2 pathogenesis, coagulopathy, and the inflammatory response.79,80
Our study highlights the potential role of MO/Mφ MLL1 in regulating coagulation/fibrinolysis, and our findings add to the evidence that CMEs play major roles in directing MO/Mφ gene expression programs to drive micro/macrovascular immunopathology.32,42,81-85 Epigenetics in translational thrombosis represents an exciting area of discovery that has the potential to uncover novel immune-based therapeutic strategies in the study of vascular immunobiology. We have identified that MLL1 is a coronavirus-inducible factor in MO/Mφ that is responsible for the expression of inflammatory and coagulopathy-related gene expression. Our study shows that MO/Mφ MLL1 induces a hypercoagulable and fibrinolytic phenotype upon coronavirus infection. These results point to MLL1 blockade as a potential therapeutic strategy to curb CAC and inflammation, and possible long-term sequelae associated with SARS-CoV-2 infection.
Acknowledgments
The authors thank Jim Morrissey (Departments of Biological Chemistry and Internal Medicine; University of Michigan, Ann Arbor, MI) for his thoughtful advice on experimental design and Lorie Gavulic for design of the visual abstract.
The authors would like to acknowledge support from the Advanced Genomics Core and Bioinformatics Core of the University of Michigan Medical School’s Biomedical Research Core Facilities and the Fredrick A. Coller Surgical Society. This work was supported by funding from the Vascular Cures Foundation (A.T.O.), the Lefkofsky Family Foundation (A.T.O.), the Elizabeth Anne Baiardi Research Fund (A.T.O.), the University of Michigan Cardiovascular Center COVID Igniter grant (A.T.O.), the National Institutes of Health/National Institute of General Medical Sciences (grant R35GM131835 to M.H.), the National Institutes of Health/National Center for Advancing Translational Sciences (grant R21TR003185 to M.H.), and the National Institutes of Health/National Heart, Lung, and Blood Institute (grants ZIAHL006267, ZIAHL006262, ZIAHL006263 to Y.K.; 5R01HL144550 to P.K.H.; 1K08HL155408-01 to A.T.O.).
Authorship
Contribution: S.B.S., W.J.M., C.O.A., M.B., N.R., Y.K., J.S.K., R.A., M.H., B.B.M., P.K.H., T.W.W., K.A.G., and A.T.O. designed research strategy; S.B.S., W.J.M., C.O.A., M.B., N.R., and A.T.O. performed research; S.B.S., W.J.M., C.O.A., W.W., N.R., R.A., M.H., B.B.M, P.K.H, K.A.G., and A.T.O. contributed new reagents/analytic tools; W.W. performed analysis of sequencing data; S.B.S., W.J.M., C.O.A., N.R., R.A., M.H., B.B.M., T.W.W., P.K.H., K.A.G., and A.T.O. analyzed data; and S.B.S. and A.T.O. wrote the manuscript.
Conflict-of-interest disclosure: M.H. is a consultant and equity holder for Veralox Therapeutics and a consultant for Cereno Scientific, which has an option to license platelet inhibitory compounds from the University of Michigan. The remaining authors declare no competing financial interests.
Correspondence: Andrea T. Obi, Department of Surgery, University of Michigan, 1500 East Medical Ctr Dr, 5364 CVC, Ann Arbor, MI 48109; e-mail: easta@med.umich.edu.
References
Author notes
Data are available on request from the corresponding author, Andrea T. Obi (easta@med.umich.edu).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![Coronavirus infection of MO/Mφs induces expression of MLL1, coagulopathy-related factors, and inflammatory cytokines. BMDMs were harvested from C57Bl6/J mice (wild-type-BMDMs) and were infected with 1 MOI of the murine coronavirus MHVA59 for the indicated times, and mRNA (A) and protein levels (B; representative blot shown [β-actin served as loading control]) of Kmt2a/MLL1 were assayed by qRT-PCR and immunoblotting, respectively. (C) mRNA levels of factors important in CAC (Plau, Plaur, and F3) were measured in infected BMDMs. (D) mRNA levels of proinflammatory cytokines identified in the inflammatory signature resulting from acute SARS-CoV-2 (IL-6 and TNFα) and the MLL1-regulated cytokine IL-1β were measured in infected BMDMs. (E-F) Protein levels of coagulopathy-related factors (E) and proinflammatory cytokines (F) were assayed by ELISA. Bar graphs represent mean values from at least n = 5 independent experiments assayed in triplicate, and individual data points represent independent experiments. Errors bars represent SE. Statistical testing was performed using Kruskal-Wallis tests with corrections for multiple comparisons. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ELISA, enzyme-linked immunosorbent assay; MOI, multiplicity of infection; qRT, quantitative reverse transcription; SE, standard error.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/7/10.1182_blood.2022015917/4/m_blood_bld-2022-015917-gr1.jpeg?Expires=1770141900&Signature=20vTa5aHNgGxykpqKdY2UhNp2Pz1r1RSDSrSc-09v59W83kYmon65tfqCyt-P6urOy-VZASrATYRcNeUnebfX73dRL~EgdXsdGFAgTmFsf14Qswahj1WRbul8A-JmWJg4VfqE~DLb2dRs3Nv6uBZbzo~1uz6ojRGCZ7S8MqY7XzwoTrulr16IyQO9y2aXOciM4MOUelbQeTmbLIB~~RNFWfgQrSQeLldTss~Z0x3iHgrEu-yO0zIG-bw1hS06jQHx-nv7RBRqd0OYFwquKXf5AYGWsegI~IIgA9opqlxHGXVmg28I269SUXfI8fwr~ZXddf2q6e7kFxaDB5777hIEQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![MLL1 regulates the basal expression of coagulopathy-related factors and proinflammatory cytokines in BMDMs. BMDMs were harvested from mice carrying a myeloid-specific deletion of MLL1 (Kmt2afl/fl Lyz2Cre+/−; denoted Cre+) and littermate controls (Kmt2afl/fl Lyz2Cre−/−; denoted Cre−). (A) mRNA levels of Kmt2a were assayed in n = 8 Cre+ and Cre− animals analyzed in triplicate. (B) Protein levels of MLL1 were assayed in BMDMs from n = 4 Cre+ and Cre− mice by immunoblotting (representative blot shown [β-actin served as loading control]). (C-D) mRNA levels of coagulopathy-related factors (C) and proinflammatory cytokines (D) were assayed. (E-F) Protein levels of coagulopathy-related factors (E) and inflammatory cytokines (F) were measured by ELISA. (G) ChIP assays were performed using antibodies specific to either H3K4me3 or nontargeting species-specific IgG. Bars graphs represent mean values from at least n = 4 independent experiments with individual data points representing independent experiments. For ChIP experiments, bar graphs represent mean ChIP intensity relative to IgG derived from n = 8 samples performed in triplicate. Statistical analysis of pairwise comparisons was performed using Mann-Whitney tests. Error bars represent SE. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ChIP, chromatin immunoprecipitation; ELISA, enzyme-linked immunosorbent assay; IgG, immunoglobulin G; SE, standard error.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/7/10.1182_blood.2022015917/4/m_blood_bld-2022-015917-gr2.jpeg?Expires=1770141900&Signature=3~NcLcHWLstxYuTSRDEUo5SxqHFBJVJRcWjG9RXo8TQxcrcpfLyuyhYWTtzcG9EFFzdrPKTQOQr-WVaNiZUUS8JCnUbLGi0woEednM6hJXKYu-2k5EJeupYXu8tAk~CX7zdOcDtie9Jd7bWRVVnmTmJRfkMYZO2MY2VlnBucmkIIyTCvNoNYw5fP-DDbssbad5qRQNajmVM~Ucim8H9Rm8ZJe69TqQ0tprkK2YijQwKAV134jHLkujJYiXmUDdX7rhfxboyboGgIyrdQDUGYH3RI1hLGZdWiyoCxwndkWEaSULddjYp6VlwYT7WNG1d5WoigCV3QsaOp9td4h46P9g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Coronavirus induces the expression of MLL1 and its associated factors in MO/Mφs in vivo and promotes a prothrombotic and profibrinolytic phenotype. C57BL6/J mice underwent intranasal inoculation of 2 × 105 plaque-forming units (pfu) MHVA59 (postinfection day 3 mice [d3; n = 8]; postinfection day 28 mice [d28; n = 9]) or PBS (denoted as sham; n = 8). Mice were sacrificed at the indicated time points and plasma and splenic MO/Mφs (a surrogate for circulating MO/Mφs) were harvested. (A) Kmt2a mRNA levels were assayed by qRT-PCR. (B-C) The mRNA levels and protein levels of coagulopathy-associated factors in splenic MO/Mφs were measured by qRT-PCR and ELISA, respectively. (D) H3K4me3 abundance at the indicated promoters was assayed by ChIP assay. (E-F) Circulating levels of PLAU and PLAUR were measured by ELISA. (G) Plasma TF protein levels was measured by ELISA. (H) Plasma PLAU activity levels were measured using a colorimetric assay in which absorbance (A405) correlates with enzyme activity level through the cleavage of a plasmin (activated by PLAU) substrate that liberates p-nitroaniline. (I) Plasma TF activity was measured using a colorimetric assay in which the activation of factor X (FXa) by TF and factor VII (TF/FVIIa) and its cleavage of a FXa-specific substrate liberates p-nitroaniline. (J) The TF activity of lysed harvested splenic MO/Mφs was measured. (K) Tail vein bleeding time was measured in infected and sham mice. (L) The number of rebleeding events during tail vein bleeding time assays was tallied. (M) Whole blood was collected from infected and sham mice by inferior vena cava puncture and anticoagulated with 3.2% sodium citrate at a ratio of 9:1 (blood to citrate). TEG was performed and R time (time to formation of clot of 2 mm thickness) was measured (left panel). Representative TEGs are presented in the right panel. (N) To determine the role of TF in hypercoagulability as assayed by a shortened R time as measured by TEG after coronavirus infection, citrated whole blood samples from either sham or infected (d3) mice was treated with either corn trypsin inhibitor (CTI; 25 μg/mL final concentration) or a mouse specific anti-TF neutralizing antibody (TFI; clone 1H1 [Genentech]; 50 μg/mL final concentration) and the resultant viscoelastic properties were analyzed using TEG. Samples that were not subjected to treatment (NT) served as controls. Bar graphs represent mean values. qRT-PCR, ELISA, and ChIP data represent experiments performed in triplicate. Error bars represent SE. Statistical analysis of data sets was performed by either Mann-Whitney U test or Kruskal-Wallis test with corrections for multiple comparisons. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ChIP, chromatin immunoprecipitation; ELISA, enzyme-linked immunosorbent assay; PBS, phosphate-buffered saline; qRT, quantitative reverse transcription; SE, standard error](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/7/10.1182_blood.2022015917/4/m_blood_bld-2022-015917-gr5.jpeg?Expires=1770141900&Signature=bMEi~B8QnnlYW9rWA2F0UTQO0CcGb4jFT43GG-P07bU3B7jvfZT0xCkBzJxWoglv9qt6r-u8RfRs6ZPpRJfkbYVhixa~MfxGoNibyTkfZyMJs-P5onyqVTEwRnML~VJxlL4RR1NvZHitDFd7PNcXsTzHDvcbY052sV7iP2SPWOmG-4bbNrdBAbbWAOg9Ud9BU2026frNyw1NPssJTVliCWBdd168IyEbqnQ1NbaI131m6RjYLUTy-qvj6oTkf1Cls3LV7IpCxifqwPEHVOvdKoH4W~bbpyMR-ZMlsnwZZl4awJ2ztZ~SJtPmSETqjbGG4auLuGH39OnyOY8NEnJtTw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal