Key Points
NTBI uptake by LSECs is the major signal for Bmp6 induction during iron overload, and Tfr1 contributes mostly under low iron conditions.
Bmp6 induction in the presence of NTBI is associated with extensive genetic reprogramming of LSECs that involves Nrf2 and Myc pathways.
Abstract
Homeostatic adaptation to systemic iron overload involves transcriptional induction of bone morphogenetic protein 6 (BMP6) in liver sinusoidal endothelial cells (LSECs). BMP6 is then secreted to activate signaling of the iron hormone hepcidin (HAMP) in neighboring hepatocytes. To explore the mechanism of iron sensing by LSECs, we generated TfrcTek-Cre mice with endothelial cell–specific ablation of transferrin receptor 1 (Tfr1). We also used control Tfrcfl/fl mice to characterize the LSEC-specific molecular responses to iron using single-cell transcriptomics. TfrcTek-Cre animals tended to have modestly increased liver iron content (LIC) compared with Tfrcfl/fl controls but expressed physiological Bmp6 and Hamp messenger RNA (mRNA). Despite a transient inability to upregulate Bmp6, they eventually respond to iron challenges with Bmp6 and Hamp induction, yet occasionally to levels slightly lower relative to LIC. High dietary iron intake triggered the accumulation of serum nontransferrin bound iron (NTBI), which significantly correlated with liver Bmp6 and Hamp mRNA levels and elicited more profound alterations in the LSEC transcriptome than holo-transferrin injection. This culminated in the robust induction of Bmp6 and other nuclear factor erythroid 2–related factor 2 (Nrf2) target genes, as well as Myc target genes involved in ribosomal biogenesis and protein synthesis. LSECs and midzonal hepatocytes were the most responsive liver cells to iron challenges and exhibited the highest expression of Bmp6 and Hamp mRNAs, respectively. Our data suggest that during systemic iron overload, LSECs internalize NTBI, which promotes oxidative stress and thereby transcriptionally induces Bmp6 via Nrf2. Tfr1 appears to contribute to iron sensing by LSECs, mostly under low iron conditions.
Introduction
Liver sinusoidal endothelial cells (LSECs) play a critical role in iron sensing by secreting bone morphogenetic protein 6 (BMP6) and BMP2, which are potent inducers of the iron regulatory hormone hepcidin.1 They can form heterodimers and bind to BMP receptors on hepatocytes to activate the SMAD signaling cascade, leading to transcription of the hepcidin-encoding HAMP gene. Hepcidin limits iron entry into the bloodstream by binding to the iron exporter ferroportin in target cells such as duodenal enterocytes and tissue macrophages, which are crucial for dietary iron absorption and iron recycling from effete red blood cells (RBCs), respectively.2 Hepcidin binding occludes ferroportin’s iron channel3 and also triggers ferroportin internalization and lysosomal degradation.4
Hepcidin expression is induced in response to elevated serum or tissue iron levels, and serves as a homeostatic adaptation to prevent systemic iron overload and ensuing complications. Increased transferrin (Tf) saturation in the blood stabilizes Tf receptor 2 (Tfr2) on hepatocytes, which in turn stimulates SMAD signaling to hepcidin.5 On the other hand, excessive tissue iron promotes induction of primarily BMP6 and to a lesser extent BMP2 in the liver.1 LSECs are the major site for hepatic BMP6 and BMP2 production, and endothelial cell (EC)–specific ablation of either Bmp66 or Bmp27,8 in mice causes systemic iron overload (hemochromatosis) due to hepcidin deficiency.
However, the mechanisms by which LSECs sense iron and respond to increased liver iron stores via BMP6 induction are poorly understood. In vitro, treatment of primary murine LSECs or LSEC-like cell lines with iron is sufficient to activate Bmp6 messenger RNA (mRNA) in a cell autonomous manner.9-11 Biochemical studies showed that the BMP6 promoter contains a binding site of the nuclear factor erythroid 2–related factor 2 (Nrf2) transcription factor and provided evidence that iron-dependent induction of BMP6 in the LSEC culture models involves Nrf2 activation by oxidative stress.9 In line with this finding, Nrf2−/− mice exhibited a blunted Bmp6 response to oral or parenteral iron challenges.9 The above data established a role of Nrf2 in iron-mediated induction of Bmp6 within LSECs, but do not offer a clue on how these cells accumulate excess iron and respond to systemic iron overload in vivo.
Most cells acquire iron from circulating Tf by Tfr1 via endocytosis.12 Thus, we hypothesized that Tf/Tfr1-mediated iron uptake may be a crucial component of the iron-sensing pathway by LSECs that results in Bmp6 induction. To address this, we generated TfrcTek-Cre mice with EC-specific ablation of the Tfr1-encoding Tfrc gene. We showed that, with minor exceptions, these mice responded to iron challenges with Bmp6 and Hamp mRNA induction; however, they tended to have relatively increased liver iron content (LIC). Iron-dependent Bmp6 and Hamp mRNA induction in both Tfrcfl/fl and TfrcTek-Cre mice positively correlated with nontransferrin bound iron (NTBI), which emerged following high dietary iron intake. Moreover, single-cell transcriptomics has shown that acute dietary iron loading promotes extensive reprogramming of LSEC gene expression in control Tfrcfl/fl mice, which is characterized by the induction of Nrf2 and Myc target genes.
Methods
Animals
Tfrcfl/fl mice were generated in-house13 and Tek-Cre transgenic mice14 (B6.Cg-Tg(Tek-Cre)1Ywa/J; JAX stock #008863) were purchased from the Jackson Laboratory (Bar Harbor, ME). Rosa26mT-mG/mT-mG reporter mice15 were kindly provided by Ernesto Schiffrin. These animals, as well as the TfrcTek-Cre and Rosa26mT-mG/+;Tfrcfl/+;Tek-Cre progeny, were housed in macrolone cages (up to 5 mice per cage, 12:12 hours light-dark cycle: 7 AM to 7 PM; at 22 ± 1°C and 60 ± 5% humidity) and were allowed ad libitum access to chow and drinking water. The mice were fed a standard diet (SD) (Teklad global 18% protein 2018, 200 parts per million iron) or, when indicated, an iron-deficient diet (IDD) (TD.80396, 2-6 parts per million iron) or a high iron diet (HID) (TD.09521, 2% carbonyl iron). Where indicated, the mice were injected IV (tail vein) with 0.9 mg ferric ammonium citrate (FAC) or 10 mg holo-Tf. At end point, animals were euthanized by CO2 inhalation. The experimental procedures were approved by the Animal Care Committee of McGill University (protocol 4966).
Biochemical assays and histology
Serum biochemistry, quantitative real-time polymerase chain reaction, immunohistochemistry, and quantification of tissue iron were performed as previously described.16-18 The details are provided in the supplemental Methods, available on the Blood website.
Single-cell transcriptomics
Liver cells were isolated from perfused anesthetized mice for single-cell RNA sequencing (scRNA-seq) analysis. Details are provided in the supplemental Methods.
Results
TfrcTek-Cre mice exhibit Tfr1 ablation in LSECs
To disrupt Tfr1 expression in LSECs, Tfrcfl/fl mice were mated with Tek-Cre transgenic animals. The resulting TfrcTek-Cre offspring were born in lower than Mendelian ratios, which is indicative of partial embryonic lethality. Moreover, some newborn pups had a pale appearance and died within a few days of birth. Approximately 25% of the expected TfrcTek-Cre mice (86/342) survived to adulthood without apparent phenotypic abnormalities. There was no sex-specific bias as the male to female ratio among the surviving animals was 1:1. Adult TfrcTek-Cre mice did not manifest anemia, but their RBCs had a modestly increased distribution width and lower mean corpuscular volume than those of Tfrcfl/fl littermates (supplemental Table 1). Additionally, RBCs from male but not female TfrcTek-Cre mice had lower mean corpuscular hemoglobin content (supplemental Table 1). There were no differences in the serum iron indices between males and females (supplemental Figure 1). These data suggest that EC-specific ablation of Tfr1 in this TfrcTek-Cre mouse model elicits strong adverse effects during embryogenesis and early postnatal period. Nevertheless, surviving mice develop normally and exhibit only a mild hematologic phenotype.
Liver sections from adult Tfrcfl/fl control and TfrcTek-Cre mice were used to validate Tfr1 ablation in LSECs using immunofluorescence. The animals were previously fed an IDD to stimulate Tfr1.13 As expected,13 Tfr1 was strongly expressed in hepatocytes of both genotypes (Figure 1A, red color). Staining for the EC-specific marker CD31 (green) revealed the LSEC lining in the sinusoids. In control livers, a Tfr1 signal was present throughout the lining (Figure 1A, left), and areas with overlapping CD31 and Tfr1 staining were visible (yellow, see rectangles 1 and 2). In contrast, the Tfr1 signal was absent in the LSEC lining of the TfrcTek-Cre mice (Figure 1A, right; see rectangles 3 and 4). Thus, the TfrcTek-Cre mouse model showed efficient Tfr1 ablation in the LSECs. We further validated this using a Rosa26mT-mG/+;Tfrcfl/+;Tek-Cre reporter mouse, where Cre recombination resulted in the replacement of red mT (membrane-targeted tandem dimer Tomato) with green mG (membrane-targeted enhanced green fluorescent protein) in the LSEC lining (Figure 1B).
Validation of the LSECs Tfr1 knockout in the TfrcTek-Cremouse model. (A) Frozen liver slices from control Tfrcfl/fl (left) or TfrcTek-Cre mice (right) were processed for immunofluorescence and stained for Tfr1 using a Cy3-labeled secondary antibody (red) and for CD31 using an Alexa488-labeled secondary antibody (green). Overlapping areas of Tfr1 and CD31 expressions are shown in yellow. Nuclei were visualized using 4′,6-diamidino-2-phenylindole (DAPI) staining (blue). Areas in the highlighted rectangles are shown at a higher magnification (bottom). Arrows indicate exclusive CD31 expression in the LSEC lining. Scale bars, 10 μm (highlighted rectangles, bottom, scale bars, 2 μm). (B) Frozen liver slices from a Rosa26mT-mG/+;Tfrcfl/+;Tek-Cre reporter mouse were processed for confocal microscopy imaging for the expression of mT (red) and mG (green); the latter emerged following Cre-mediated recombination. The arrows indicate the LSEC lining. Scale bars, 10 μm.
Validation of the LSECs Tfr1 knockout in the TfrcTek-Cremouse model. (A) Frozen liver slices from control Tfrcfl/fl (left) or TfrcTek-Cre mice (right) were processed for immunofluorescence and stained for Tfr1 using a Cy3-labeled secondary antibody (red) and for CD31 using an Alexa488-labeled secondary antibody (green). Overlapping areas of Tfr1 and CD31 expressions are shown in yellow. Nuclei were visualized using 4′,6-diamidino-2-phenylindole (DAPI) staining (blue). Areas in the highlighted rectangles are shown at a higher magnification (bottom). Arrows indicate exclusive CD31 expression in the LSEC lining. Scale bars, 10 μm (highlighted rectangles, bottom, scale bars, 2 μm). (B) Frozen liver slices from a Rosa26mT-mG/+;Tfrcfl/+;Tek-Cre reporter mouse were processed for confocal microscopy imaging for the expression of mT (red) and mG (green); the latter emerged following Cre-mediated recombination. The arrows indicate the LSEC lining. Scale bars, 10 μm.
TfrcTek-Cre mice express physiological levels of Bmp6 and Hamp mRNAs and induce them in response to iron
We explored the effect of EC-specific Tfr1 ablation on the sensing of high dietary iron by LSECs (Figure 2A). The first group of Tfrcfl/fl and TfrcTek-Cre mice were fed a SD and served as controls. A second group of animals were switched from SD to a HID for 18 hours. This dietary manipulation was expected to rapidly increase Tf saturation leading to gradual NTBI accumulation.13 For acute exposure to NTBI, mice in a third group received an IV injection with FAC and euthanized after 5 hours, an optimal time point inferred from a preliminary kinetics experiment (supplemental Figure 2).
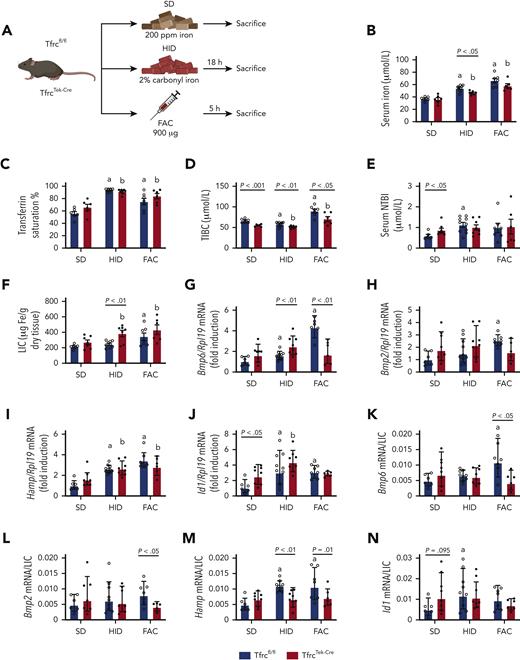
TfrcTek-Cremice express physiological levels of Bmp6 and Hamp mRNAs and induce them in response to high dietary iron, but fail to induce Bmp6 mRNA following FAC injection. (A) Schematic experimental outline. All mice were 7- to 8-week-old males (n = 6-10 per group). At end point, the animals were euthanized, serum was prepared, and livers were harvested for biochemical analysis. Serum iron (B); Tf saturation (C); total iron binding capacity (TIBC) (D); NTBI (E); LIC (F); quantitative polymerase chain reaction analysis of mRNAs encoding liver Bmp6, Bmp2, hepcidin (Hamp), and Id1, respectively (G-J); and mRNA/LIC ratios for Bmp6, Bmp2, Hamp and Id1, respectively (K-N). Serum data (B-E) and LIC (F) are represented as mean ± standard error of the mean, and gene expression data (G-N) are represented as geometric mean ± geometric standard deviation. Statistical differences (P < .05) were determined using Student t test on the original data (B-E) or log-transformed gene expression data (G-N). Statistically significant differences between Tfrcfl/fl and TfrcTek-Cre mice on the SD are represented as a and b, respectively. Illustration of panel A was performed using BioRender.com. Fe, iron; ppm, parts per million.
TfrcTek-Cremice express physiological levels of Bmp6 and Hamp mRNAs and induce them in response to high dietary iron, but fail to induce Bmp6 mRNA following FAC injection. (A) Schematic experimental outline. All mice were 7- to 8-week-old males (n = 6-10 per group). At end point, the animals were euthanized, serum was prepared, and livers were harvested for biochemical analysis. Serum iron (B); Tf saturation (C); total iron binding capacity (TIBC) (D); NTBI (E); LIC (F); quantitative polymerase chain reaction analysis of mRNAs encoding liver Bmp6, Bmp2, hepcidin (Hamp), and Id1, respectively (G-J); and mRNA/LIC ratios for Bmp6, Bmp2, Hamp and Id1, respectively (K-N). Serum data (B-E) and LIC (F) are represented as mean ± standard error of the mean, and gene expression data (G-N) are represented as geometric mean ± geometric standard deviation. Statistical differences (P < .05) were determined using Student t test on the original data (B-E) or log-transformed gene expression data (G-N). Statistically significant differences between Tfrcfl/fl and TfrcTek-Cre mice on the SD are represented as a and b, respectively. Illustration of panel A was performed using BioRender.com. Fe, iron; ppm, parts per million.
Intake of HID increased serum iron, Tf saturation, and NTBI in both Tfrcfl/fl and TfrcTek-Cre mice within 18 hours (Figure 2B-E). Similar results were obtained with FAC injection; however, the NTBI increase did not reach statistical significance, presumably due to rapid clearance by liver parenchymal cells, as previously shown.19 TfrcTek-Cre mice tended to have higher LIC in all experimental settings (Figure 2F), but their iron content in spleen, kidney, and heart was physiological (supplemental Figure 3). Expression of the mRNAs encoding Bmp6, Bmp2, and the BMP target hepcidin (Hamp) did not significantly differ in the livers of Tfrcfl/fl and TfrcTek-Cre mice on SD (Figure 2G-H), and this persisted after normalization to LIC (Figure 2K-M). Expression of the BMP target Id and Id1/LIC ratios were higher in TfrcTek-Cre mice (Figure 2J and N, respectively). As expected, HID intake induced Bmp6, Hamp, and Id1 mRNA expression in both genotypes; however, the Hamp/LIC ratio was lower in TfrcTek-Cre mice. Surprisingly, FAC injection promoted Bmp6 mRNA induction only in Tfrcfl/fl mice but not in TfrcTek-Cre mice (Figure 2G). It also resulted in Hamp mRNA upregulation in both genotypes (Figure 2I), possibly because of the contribution of the inflammatory pathway, as FAC induced the expression of the inflammatory marker Socs3 (supplemental Figure 4). Taken together, these data suggest that the response of Tfr1-deficient LSECs to high dietary iron levels is intact. However, Tfr1 appeared to be critical for Bmp6 induction following FAC injection, at least within the 5 hours experimental time frame.
Iron-restricted TfrcTek-Cre mice have increased LIC, express relatively lower levels of Bmp6 and Hamp mRNAs, and respond to dietary iron or holo-Tf injection
In another experiment, we addressed the responses of Tfrcfl/fl and TfrcTek-Cre mice to a more physiologically relevant dietary iron intake or holo-Tf injection (Figure 3A). All animals were rendered relatively iron deficient by feeding an IDD for 1 week and then divided into 3 experimental groups. In the first group, the mice remained on the IDD and served as controls. In the second and third groups, the mice were either switched from IDD to SD or injected IV with holo-Tf, and then euthanized (together with controls) after 18 hours.
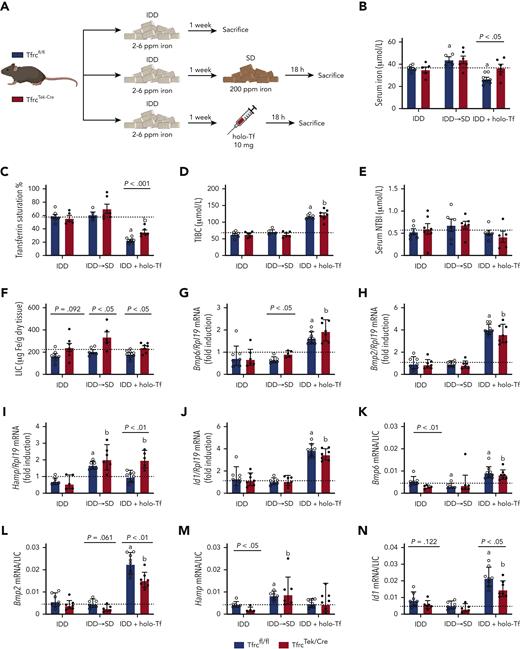
TfrcTek-Cremice on IDD have increased LIC, express relatively low Bmp6 and Hamp mRNA levels, and induce them in response to dietary iron or holo-Tf. (A) Schematic experimental outline. All mice were 7- to 8-week-old males (n = 6-10 per group). At end point, the animals were euthanized; serum was prepared, and livers were harvested for biochemical analysis. Serum iron (B); Tf saturation (C); TIBC (D); NTBI (E); LIC (F); qPCR analysis of mRNAs encoding liver Bmp6, Bmp2, Hamp, and Id1, respectively (G-J); and mRNA/LIC ratios for Bmp6, Bmp2, Hamp, and Id1, respectively (K-N). Serum data (B-E) and LIC (F) are represented as mean ± standard error of the mean and gene expression data (G-N) are represented as geometric mean ± geometric standard deviation. Statistical differences (P < .05) were determined using Student t test on original data (B-E) or log-transformed gene expression data (G-N). Statistically significant differences from Tfrcfl/fl and TfrcTek-Cre mice on SD are represented as a or b, respectively. Dotted lines indicate the average values obtained from age-matched male Tfrcfl/fl mice on SD (n = 6). Illustration of panel A was made with BioRender.com.
TfrcTek-Cremice on IDD have increased LIC, express relatively low Bmp6 and Hamp mRNA levels, and induce them in response to dietary iron or holo-Tf. (A) Schematic experimental outline. All mice were 7- to 8-week-old males (n = 6-10 per group). At end point, the animals were euthanized; serum was prepared, and livers were harvested for biochemical analysis. Serum iron (B); Tf saturation (C); TIBC (D); NTBI (E); LIC (F); qPCR analysis of mRNAs encoding liver Bmp6, Bmp2, Hamp, and Id1, respectively (G-J); and mRNA/LIC ratios for Bmp6, Bmp2, Hamp, and Id1, respectively (K-N). Serum data (B-E) and LIC (F) are represented as mean ± standard error of the mean and gene expression data (G-N) are represented as geometric mean ± geometric standard deviation. Statistical differences (P < .05) were determined using Student t test on original data (B-E) or log-transformed gene expression data (G-N). Statistically significant differences from Tfrcfl/fl and TfrcTek-Cre mice on SD are represented as a or b, respectively. Dotted lines indicate the average values obtained from age-matched male Tfrcfl/fl mice on SD (n = 6). Illustration of panel A was made with BioRender.com.
Under iron-restricted conditions, TfrcTek-Cre mice exhibited modestly but significantly elevated LIC and reduced Bmp6/LIC and Hamp/LIC ratios compared with control Tfrcfl/fl littermates (Figure 3F,K,M). Switching from IDD to SD tended to increase serum iron, Tf saturation, and NTBI in both genotypes, whereas holo-Tf injection did not affect serum iron at the end point but increased total iron binding capacity and decreased Tf saturation without alterations in NTBI (Figure 3B-E). Switching to SD did not alter the expression of Bmp6, Bmp2, or Id1 mRNAs but promoted Hamp mRNA induction to levels appropriate relative to LIC in both Tfrcfl/fl and TfrcTek-Cre mice (Figure 3G-N). Hamp mRNA was also induced following holo-Tf injection and the response was more potent in TfrcTek-Cre mice (Figure 3I). The Hamp/LIC ratio was lower at baseline (IDD) in TfrcTek-Cre mice (Figure 3M), although the Hamp mRNA expression was at the control level (Figure 3I). Surprisingly, holo-Tf injection promoted the induction of Bmp6, Bmp2, and Id1 mRNAs expression in both Tfrcfl/fl and TfrcTek-Cre mice (Figure 3G-N). Dietary iron manipulation or holo-Tf injections did not affect the expression of Socs3 mRNA (supplemental Figure 4).
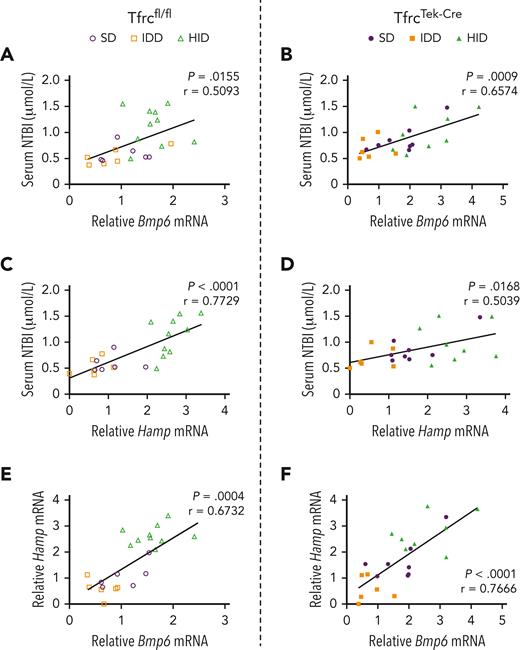
The expression of Bmp6 and Hamp mRNAs was positively and significantly correlated with NTBI (Figure 4), as well as with Tf saturation and LIC (supplemental Figure 5) in both Tfrcfl/fl and TfrcTek-Cre mice. As expected, the correlation between Bmp6 and Hamp mRNA levels was also significant (Figure 4E-F). The correlations between NTBI and Tf saturation or LIC were significant only in the control Tfrcfl/fl mice but not in TfrcTek-Cre mice (supplemental Figure 5I-L). Together, the above data suggest that Tfr1 ablation in LSECs of TfrcTek-Cre mice is associated with a modest increase in LIC, but only minimally affects the capacity of these animals to induce Bmp6 in response to iron challenges.
Expression of Bmp6 and Hamp mRNAs positively correlates with serum NTBI in both Tfrcfl/fland TfrcTek-Cremice. Pearson correlation analysis of NTBI vs Bmp6 mRNA (A-B); NTBI vs Hamp mRNA (C-D); and Hamp mRNA vs Bmp6 mRNA (E-F). Data are from the experiments shown in Figures 2 and 3 with Tfrcfl/fl and TfrcTek-Cre mice kept on a SD, exposed to a 2% carbonyl HID for 18 hours, or placed on an IDD for 1 week. P values and Pearson r coefficients are provided for each graph.
Expression of Bmp6 and Hamp mRNAs positively correlates with serum NTBI in both Tfrcfl/fland TfrcTek-Cremice. Pearson correlation analysis of NTBI vs Bmp6 mRNA (A-B); NTBI vs Hamp mRNA (C-D); and Hamp mRNA vs Bmp6 mRNA (E-F). Data are from the experiments shown in Figures 2 and 3 with Tfrcfl/fl and TfrcTek-Cre mice kept on a SD, exposed to a 2% carbonyl HID for 18 hours, or placed on an IDD for 1 week. P values and Pearson r coefficients are provided for each graph.
Single-cell transcriptomic profiles in the mouse liver following holo-Tf injection or acute dietary iron loading
We determined the effect of holo-Tf injection and acute dietary iron loading on the transcriptome of liver cells using scRNA-seq (Figure 5A). Three Tfrcfl/fl mice were fed an IDD for 1 week, one of which was used as the control. The remaining mice were either injected with holo-Tf or switched to HID for 18 hours. At end point, the livers were dissociated, and single-cell suspensions were used for scRNA-seq analysis. This produced good quality data sets (supplemental Figure 6A-B), with 2070 cells in the control (IDD) group, 6505 cells in the holo-Tf (IDD + holo-Tf) group, and 3134 cells in the HID (IDD to HID) group (Figure 5B). Dimensionality reduction followed by clustering and differential gene expression analysis allowed us to annotate 9 distinct cell types: hepatocytes (2 types), macrophages/monocytes, ECs, B lymphocytes, stellate cells, T lymphocytes, dendritic cells, and neutrophils (Figure 5B-C; supplemental Figure 5C). These cell types expressed known gene markers (Figure 5C) and were found in variable proportions in each data set (Figure 5D).
scRNA-seq identifies ECs and midzonal hepatocytes as the most responsive cell types to acute dietary iron loading or holo-Tf injection. (A) Schematic experimental outline of scRNA-seq experiment. All mice were 7- to 8-week-old male Tfrcfl/fl mice (n = 1 per group). (B) UMAP (uniform manifold approximation and projection) plot of the most common liver cell types. (C) Dot plot annotation based on differentially expressed genes and previously published cell type markers. (D) Relative proportion of each liver cell population obtained from the experimental settings. (E) Cell type prioritization (lollipop plot) based on Augur-calculated area under the curve (AUC) scores for each comparison (IDD vs IDD + holo-Tf, IDD vs IDD to HID, and IDD to HID vs IDD + holo-Tf). (F) Heat maps with log2 (FC) of gene set variation analysis score for iron-related biological pathways from the Molecular Signatures Database across cell types identified by scRNA-seq, highlighting the most responsive cell types to different iron manipulations. BCs, B cells; DCs, dendritic cells; DEGs, differentially expressed genes; ECs, endothelial cells; GO, gene ontology.
scRNA-seq identifies ECs and midzonal hepatocytes as the most responsive cell types to acute dietary iron loading or holo-Tf injection. (A) Schematic experimental outline of scRNA-seq experiment. All mice were 7- to 8-week-old male Tfrcfl/fl mice (n = 1 per group). (B) UMAP (uniform manifold approximation and projection) plot of the most common liver cell types. (C) Dot plot annotation based on differentially expressed genes and previously published cell type markers. (D) Relative proportion of each liver cell population obtained from the experimental settings. (E) Cell type prioritization (lollipop plot) based on Augur-calculated area under the curve (AUC) scores for each comparison (IDD vs IDD + holo-Tf, IDD vs IDD to HID, and IDD to HID vs IDD + holo-Tf). (F) Heat maps with log2 (FC) of gene set variation analysis score for iron-related biological pathways from the Molecular Signatures Database across cell types identified by scRNA-seq, highlighting the most responsive cell types to different iron manipulations. BCs, B cells; DCs, dendritic cells; DEGs, differentially expressed genes; ECs, endothelial cells; GO, gene ontology.
The effects of holo-Tf and dietary iron on liver cell transcriptomes were first investigated using a machine learning algorithm (Augur20) implementing a random forest classifier to detect the most affected cell types. Compared with the control, holo-Tf injection and HID intake had the greatest impact on ECs, followed by stellate cells (Figure 5E). However, when comparing HID to holo-Tf treatments, midzonal hepatocytes were the most divergent, followed by B lymphocytes. Cell type–specific differential analysis between the groups showed significant gene expression changes in all cell types (supplemental Figure 7). These transcriptomic variations were associated with the differential pathway enrichment obtained from the Molecular Signatures Database (supplemental Figure 8A). Analysis of the iron pathways from this database revealed that both holo-Tf and dietary iron increased the expression of iron-related gene sets (Figure 5F; supplemental Figure 5D). Notably, these changes were more pronounced in the group switched to HID than in the holo-Tf–treated group. Across the different cell types, ECs and midzonal hepatocytes were the most responsive to the experimental treatments. Interestingly, the observed iron effects in both cell types matched with increases in reactive oxygen species (ROS)–associated pathways, which were also observed in macrophages/monocytes and dendritic cells (supplemental Figure 8B).
Iron-induced genetic reprogramming in LSECs and midzonal hepatocytes
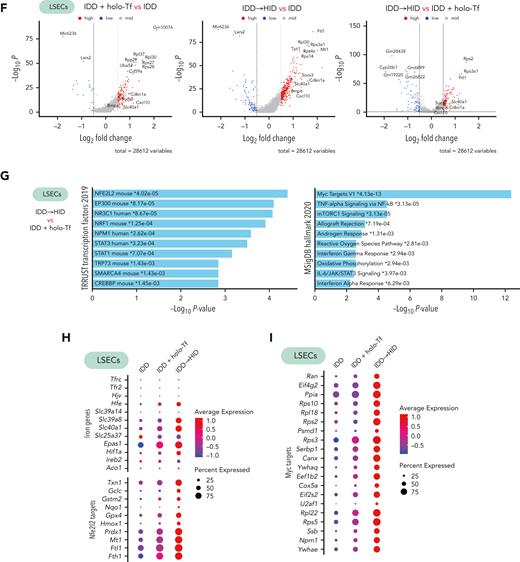
ECs line hepatic arteries, veins, and sinusoids (Figure 6A). Subclustering of ECs from our scRNA-seq data sets defined 3 subtypes expressing specific arterial (periportal), venous (pericentral), and sinusoidal (midzonal) marker genes (Figure 6B-C), in line with previous data.21 The sinusoidal subtype (LSECs) represented most of the detected ECs (Figure 6D) and expressed genes involved in Bmp and Tgf signaling, as reported22 (Figure 6E). Among the LSECs, Bmp6 and Bmp2 were primarily responsive to acute dietary iron loading (HID vs control: Bmp6, P = 6.8e-23, log2 fold change [FC] = 0.46; Bmp2, P = 1.3e-32, log2[FC] = 0.50) and less responsive to holo-Tf (holo-Tf vs control: Bmp6, P > .05; Bmp2, P = 1.2e-19, log2[FC] = 0.37). Thus, we focused on the transcriptomic impact of HID intake or holo-Tf injection on LSECs, which triggered a significant upregulation of genes compared to the control group (Figure 6F; supplemental Table 2). When comparing the transcriptome of LSECs between the HID and holo-Tf groups (Figure 6F), we found significant enrichment of Nfe2l2 (Nrf2) and Myc target genes (Figure 6G). Correlating with the increased expression of iron-related genes in the HID group, LSECs showed greater expression of genes related to the Nrf2 and Myc pathways compared with control and holo-Tf groups (Figure 6H-I). Among the iron-related genes, HID intake strongly induced the expression of Slc39a8, Slc40a1, Fth, and Ftl, encoding the metal-ion (and NTBI) transporter Zip8, the iron exporter ferroportin, and the H- and L-subunits of the iron storage protein ferritin, respectively (Figure 6H).
Acute dietary iron loading triggers activation of Nfe2l2 (Nrf2) and Myc target genes in capillary LSECs. (A) Schematic representation of the liver cell architecture. (B-D) The portal and central veins, arteries, and sinusoids of the liver contain distinct EC subtypes as identified by scRNA-seq (B, UMAP plot), which can be annotated based on differentially expressed genes (C, dot plot); with their relative cell subtype proportion in each experimental setting (D). Expression of genes involved in Bmp and Tgf signaling in EC subtypes in each experimental condition (E, dot plot) and DEGs in capillary ECs between diet conditions (F, volcano plot). (G-I) DEGs in capillary LSECs between IDD + holo-Tf and IDD to HID were enriched in the Nfe2l2 and Myc pathways (G, EnrichR) and are also represented by dot plots (H-I). Illustration of panel A was made with BioRender.com. Exp., expression.
Acute dietary iron loading triggers activation of Nfe2l2 (Nrf2) and Myc target genes in capillary LSECs. (A) Schematic representation of the liver cell architecture. (B-D) The portal and central veins, arteries, and sinusoids of the liver contain distinct EC subtypes as identified by scRNA-seq (B, UMAP plot), which can be annotated based on differentially expressed genes (C, dot plot); with their relative cell subtype proportion in each experimental setting (D). Expression of genes involved in Bmp and Tgf signaling in EC subtypes in each experimental condition (E, dot plot) and DEGs in capillary ECs between diet conditions (F, volcano plot). (G-I) DEGs in capillary LSECs between IDD + holo-Tf and IDD to HID were enriched in the Nfe2l2 and Myc pathways (G, EnrichR) and are also represented by dot plots (H-I). Illustration of panel A was made with BioRender.com. Exp., expression.
LSECs-derived BMPs activate the SMAD signaling cascade in hepatocytes, which ultimately leads to transcriptional induction of Hamp. Two clusters of hepatocytes were detected in the scRNA-seq data sets (Figure 7A). They manifested gene expression differences consistent with the transcriptomic patterns associated with liver zonation23,24 (Figure 7B). Pericentral and periportal hepatocytes tended to cluster near each other, possibly due to iron treatment conditions, and were thus grouped into “other hepatocytes.” Midzonal hepatocytes represented a smaller fraction (Figure 7C) but exhibited the greatest response to holo-Tf or HID, notably with respect to Hamp expression (Figure 7D). Moreover, gene set variation analysis showed higher expression of the Gene Ontology iron binding pathway in midzonal hepatocytes, and more importantly in the HID group (Figure 7E). Differential gene expression analysis of iron manipulation in midzonal hepatocytes revealed greater transcriptomic responses in the HID group (Figure 7F), with upregulation of Hamp, Crot, Gpx4, and Gpx1. Pathway analysis showed significant enrichment of Nrf2 target genes as well as genes involved in ROS and oxidative phosphorylation metabolism (Figure 7G-H; supplemental Table 3).
Acute dietary iron loading upregulates Nfe2l2 (Nrf2), ROS, and oxidative phosphorylation metabolic genes in midzonal hepatocytes. (A-B) Midzonal hepatocytes are separated from other hepatocyte populations identified by scRNA-seq (A, UMAP plot) and show a specific gene expression profile (B, dot plot). (C) Relative proportions of midzonal and other hepatocyte subtypes in each experimental condition. (D-E) Expression of genes involved in iron signaling (D, dot plot) and gene set variation analysis score for the GO iron ion binding pathways (E, ridge plot) in midzonal and other hepatocyte subtypes under each experimental condition. (F) Volcano plot with DEGs in midzonal hepatocytes under each experimental condition. (G-H) DEGs in midzonal hepatocytes were enriched in the Nfe2l2, ROS, and oxidative phosphorylation (OXPHOS) pathways (G, EnrichR), also represented by dot plots (H).
Acute dietary iron loading upregulates Nfe2l2 (Nrf2), ROS, and oxidative phosphorylation metabolic genes in midzonal hepatocytes. (A-B) Midzonal hepatocytes are separated from other hepatocyte populations identified by scRNA-seq (A, UMAP plot) and show a specific gene expression profile (B, dot plot). (C) Relative proportions of midzonal and other hepatocyte subtypes in each experimental condition. (D-E) Expression of genes involved in iron signaling (D, dot plot) and gene set variation analysis score for the GO iron ion binding pathways (E, ridge plot) in midzonal and other hepatocyte subtypes under each experimental condition. (F) Volcano plot with DEGs in midzonal hepatocytes under each experimental condition. (G-H) DEGs in midzonal hepatocytes were enriched in the Nfe2l2, ROS, and oxidative phosphorylation (OXPHOS) pathways (G, EnrichR), also represented by dot plots (H).
Collectively, the single-cell transcriptomic data validated the crucial roles of LSECs in iron sensing via Bmp6 (and Bmp2) and of midzonal hepatocytes in Hamp induction. Moreover, they demonstrated that acute dietary iron loading triggers more pronounced genetic responses in LSECs than holo-Tf injection, which appears to involve the Nrf2- and Myc-dependent pathways.
Discussion
LSECs respond to systemic iron overload by transcriptional induction of BMP6, which in turn activates signaling to hepcidin in neighboring hepatocytes.6 We hypothesized that iron sensing by LSECs may require uptake of Tf-bound iron via Tfr1. To address this, we used an established Tek-Cre mouse strain14 for EC-specific disruption of Tfr1 in Tfrcfl/fl mice.13 The resulting TfrcTek-Cre mice exhibited partial embryonic and neonatal lethality. Considering that Tek-Cre mice manifest variable degrees of aberrant Cre recombinase expression in hematopoietic lineages25 and that Tfr1 is essential during erythropoiesis,26 this phenotype is likely related to Tfr1 ablation in erythroid cells. It should be noted that adult mice carrying the Tek-Cre1Ywa allele (similar to the TfrcTek-Cre animals used in this study) appeared to express only a small number of Cre-positive blood cells compared with other Tek-Cre mice.25 This is consistent with the fact that the surviving adult TfrcTek-Cre mice did not develop anemia and only manifested modestly altered hematologic indices, such as distribution width, mean corpuscular volume, and mean corpuscular hemoglobin (supplemental Table 1). Efficient disruption of Tfr1 in LSECs from TfrcTek-Cre mice is shown in Figure 1.
The data in Figures 2 and 3 demonstrate that TfrcTek-Cre mice responded efficiently to dietary iron intake by inducing Bmp6 and downstream Hamp expression, despite Tfr1 ablation in LSECs. Although these homeostatic responses were largely preserved, we noted that TfrcTek-Cre animals tended to have modestly increased LIC, whereas in some instances, the iron-dependent upregulation of Bmp6 and/or Hamp mRNAs was relatively reduced when adjusted for LIC. This was more evident in the iron-restricted TfrcTek-Cre mice. Similar results were recently reported in TfrcStab2-Cre mice, another model of Tfr1 disruption in LSECs.27 Together, these data suggest a minor contribution of Tf-bound iron and Tfr1 in the iron-sensing pathway that gives rise to Bmp6 induction in LSECs. Further support for this conclusion is provided by the scRNA-seq data showing extensive reprogramming of iron pathways in LSECs by holo-Tf (Figure 5E-F), with minor Bmp6 induction compared to that achieved by acute dietary iron loading (Figure 6E).
Bmp6 and Hamp mRNA expression positively correlated with LIC, Tf saturation, and NTBI levels in both TfrcTek-Cre and control Tfrcfl/fl mice (Figure 4; supplemental Figure 5). The positive correlation between Bmp6 mRNA and LIC is in line with earlier findings.28 Nevertheless, LIC is unlikely to be the driver for Bmp6 induction in LSECs for following reasons: first, ablation of the NTBI transporter Zip14 from Hjv−/− mice, a model of hemochromatosis, prevented hepatocellular iron overload without compromising appropriate Bmp6 mRNA induction;29 second, FpnTek-Cre mice (bearing EC-specific disruption of ferroportin) were anemic and developed iron overload in Kupffer cells and hepatocytes but failed to induce Bmp6 mRNA;30 and third, exposure of primary LSECs or LSEC-like cell lines to iron was sufficient to induce Bmp6.9-11,27
In several publications, an acute increase in Tf saturation directly activated hepcidin in hepatocytes without altering Bmp6 mRNA,13,28,30 likely via Tfr2 stabilization.5 Thus, the induction of Bmp6 following holo-Tf injection in both TfrcTek-Cre and control Tfrcfl/fl mice (Figure 3G) was unexpected, even though variable responses in control animals may be related to differences in experimental design. IV injection of 10 mg holo-Tf led to an input of 250 μM extra iron into the bloodstream, corresponding to a transient but dramatic elevation of total serum iron levels by approximately sevenfold. Excess iron was cleared after 18 hours (Figure 3B), and presumably a significant fraction was taken up by liver cells; as indicated by our scRNA-Seq data which showed prominent responses of macrophages/monocytes and stellate cells to holo-Tf injection (Figure 5F). We observed that holo-Tf also upregulated the Bmp2 mRNA levels (Figure 3H).
The quantitatively similar induction of Bmp6 and Bmp2 mRNA in both genotypes (Figure 3G-H) argues against a direct effect of Tfr1-mediated uptake of excess Tf, because LSECs of TfrcTek-Cre mice are Tfr1 deficient. Nevertheless, the contribution of alternative uptake of Tf-bound iron by Tfr2 cannot be excluded, especially considering that Tfrc and Tfr2 mRNAs are expressed at comparable levels in liver ECs (supplemental Figure 6D). Alternatively, iron-loaded Kupffer and/or stellate cells can secrete activating signals. A potential candidate is ferritin, which can be released with its iron content by extracellular vesicles from iron-loaded cells31 and can activate Bmp6 expression when injected into mice.32 Moreover, ferritin can also induce Bmp6 mRNA in cultured primary LSECs.27 Conceivably, uptake of iron-rich ferritin may offer a backup mechanism for iron sensing by LSECs.
The data from the holo-Tf injection experiment in Figure 3 argue against an iron-sensing function of LSECs Tfr1. However, Figure 2G shows that control Tfrcfl/fl, but not TfrcTek-Cre mice, can induce Bmp6 following FAC injection, which implies a critical role for Tfr1. Although the aim of FAC administration was to increase serum NTBI, this procedure also increased Tf saturation. In fact, at the 5-hour end point, only high Tf saturation was sustained, and most NTBI was cleared. Thus, Bmp6 induction in Tfrcfl/fl mice was apparently due to the uptake of Tf-bound iron by LSECs Tfr1. Conversely, the failure of TfrcTek-Cre mice to induce Bmp6 could be due to insufficient exposure of Tfr1-deficient LSECs to a sustained NTBI threshold.
The seemingly contradictory data from the holo-Tf and FAC injection experiments are consistent with a model in which LSECs initially respond to an iron challenge by taking up Tf-bound iron via Tfr1, which provides an early signal for Bmp6 induction. Conceivably, once Tf saturation increases, the uptake of emerging NTBI by LSECs becomes a dominant late stimulus for Bmp6 induction, which could be enhanced by factors secreted from surrounding cells. As iron overload develops, expression of Tfr1 in LSECs and other liver cells is expected to be suppressed by post-transcriptional mechanisms.12
Our data suggest that NTBI is a major driver of prolonged iron-dependent Bmp6 induction under iron overload. It was detectable under experimental settings of acute dietary iron loading but not following holo-Tf injection. A notable difference in the genetic responses of LSECs to HID intake vs holo-Tf injection (reflecting NTBI) was the activation of a battery of Nrf2 target genes, including Bmp6 (Figure 6H). This indicates oxidative stress33 and activation of iron pathways34 and is also consistent with the known role of NTBI as an inducer of oxidative stress in ECs.35 The strong induction of Slc39a8 mRNA by acute iron loading corroborates earlier biochemical data in cell lines36 and makes Zip8 a good candidate for NTBI uptake by LSECs. This response may not be homeostatic at the cellular level but could contribute to systemic iron homeostasis by upregulating Bmp6 and Hamp. In contrast, the induction of Slc40a1, Fth, and Ftl in LSECs following acute dietary iron loading may contribute to the resolution of iron-induced oxidative stress by promoting iron efflux via ferroportin or iron storage within ferritin.
Our findings support the previously established role of Nrf2 as a Bmp6 regulator.9 Moreover, they imply that the Nrf2 pathway is not the single contributor to Bmp6 induction, as dietary iron triggers further molecular responses. The most striking finding was the induction of Myc target genes (Figure 6G), primarily encoding ribosomal proteins and translation factors (Figure 6I). Considering that Myc regulates ribosomal biogenesis and protein synthesis,37 this finding indicates stimulatory effects of iron on global mRNA translation in LSECs.
The single-cell transcriptomic data revealed the landscape of gene expression profiles in all liver cell types following acute dietary iron loading or holo-Tf injection. ECs were the primary but not the only responders to iron challenges, and iron pathways were generally more sensitive to dietary iron than holo-Tf (Figure 5F). Nevertheless, it should be emphasized that a single bolus of exogenous holo-Tf was cleared within the time frame of the experiment (Figure 3B), whereas dietary iron loading (and NTBI formation) was continuous.
Midzonal hepatocytes responded to dietary iron by inducing Hamp (Figure 7D) and by general genetic reprogramming of the iron, Nrf2, ROS, and oxidative phosphorylation pathways (Figures 5F and 7G-H). It appears that these cells are the primary producers of hepcidin and are the major regulators of systemic iron homeostasis. The zonation of Hamp mRNA in midzonal hepatocytes is consistent with earlier data.23,24
In conclusion, we demonstrated that endothelial Tfr1 contributes to the early iron-dependent induction of Bmp6 by LSECs, especially under low iron conditions. Together with previous studies, our data suggest that under iron overload, LSECs mount transcriptional Bmp6 induction in response to NTBI via an Nrf2-dependent oxidative stress mechanism. Our single-cell transcriptomic analysis revealed that Zip8 is a potential NTBI transporter. Furthermore, it identified the contribution of additional pathways, such as Myc-dependent ribosomal biogenesis and protein synthesis to NTBI-induced activation of Bmp6.
Acknowledgments
This work was funded by a grant from the Canadian Institutes of Health Research (PJT-159730). This work was supported by fellowships from the Natural Sciences and Engineering Research Council of Canada and Fonds de recherche du Québec – Santé (E.C.), and by funds from the Vision Research Health Network and Single Cell Academy (G.C.).
Authorship
Contribution: E.C., C.F., and J.P. performed the research and analyzed the data; V.L. and V.-P.L. performed and supervised the generation of single-cell RNA sequencing libraries; G.C. performed the bioinformatics analysis, produced figures related to single-cell RNA sequencing analysis, and wrote part of “Results” and “Methods”; J.-S.J. supervised the single-cell RNA sequencing experiments and data analysis; and K.P. designed and supervised the study and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kostas Pantopoulos, Lady Davis Institute for Medical Research and Department of Medicine, McGill University, 3999 Cote Ste-Catherine Rd, Montreal, QC H3T 1E2, Canada; e-mail: kostas.pantopoulos@mcgill.ca.
References
Author notes
Raw and normalized data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE215324).
Data are available on request from the corresponding author, Kostas Pantopoulos (kostas.pantopoulos@mcgill.ca).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal