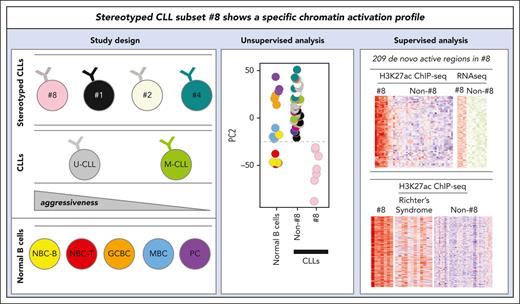
Key Points
Stereotyped subset 8, an aggressive entity associated with Richter transformation, is characterized by a unique chromatin activation profile.
A set of 209 de novo active regions in subset 8 are associated with 113 overexpressed genes, potentially related to Richter transformation.
Abstract
The chromatin activation landscape of chronic lymphocytic leukemia (CLL) with stereotyped B-cell receptor immunoglobulin is currently unknown. In this study, we report the results of a whole-genome chromatin profiling of histone 3 lysine 27 acetylation of 22 CLLs from major subsets, which were compared against nonstereotyped CLLs and normal B-cell subpopulations. Although subsets 1, 2, and 4 did not differ much from their nonstereotyped CLL counterparts, subset 8 displayed a remarkably distinct chromatin activation profile. In particular, we identified 209 de novo active regulatory elements in this subset, which showed similar patterns with U-CLLs undergoing Richter transformation. These regions were enriched for binding sites of 9 overexpressed transcription factors. In 78 of 209 regions, we identified 113 candidate overexpressed target genes, 11 regions being associated with more than 2 adjacent genes. These included blocks of up to 7 genes, suggesting local coupregulation within the same genome compartment. Our findings further underscore the uniqueness of subset 8 CLL, notable for the highest risk of Richter’s transformation among all CLLs and provide additional clues to decipher the molecular basis of its clinical behavior.
Introduction
Chronic lymphocytic leukemia (CLL) is the most common leukemia in the western world and shows a wide range of biological features with clinical associations.1 A key feature of CLL pathogenesis is the B-cell receptor immunoglobulin (BcR-IG), which represents a critical disease driver providing survival and proliferation signals. The importance of the BcR-IG is highlighted by the identification of 2 major CLL molecular subtypes based on the somatic hypermutation (SHM) status of IGHV genes. Cases displaying no or limited SHM (“unmutated” CLL, U-CLL) generally show a more aggressive clinical course compared with those with a significant SHM burden (“mutated” CLL, M-CLL).2 Another remarkable aspect underlining the significance of the BcR in CLL concerns the identification of subsets of patients expressing (quasi)identical, stereotyped BcR-IG sequences,3 strongly supporting a role for antigen selection in the natural history of CLL. Four major stereotyped subsets in CLL, that is, 1, 2, 4, and 8, which cumulatively account for ∼7% of all CLLs, mirror the heterogeneous nature of CLL as a whole. Also, these stereotyped subsets represent distinct disease variants with consistent clinico-biological features, such as different frequencies of genetic lesions (SF3B1 mutations in 2 or trisomy 12 in 84) and different outcomes.5,6
Besides immunogenetics and genetics, epigenetic studies focused on DNA methylation have further identified differential features in certain stereotyped subsets, for example, subset 2 belongs to the intermediate epigenetic subgroup7 and subset 8 shows a specific DNA methylation signature.8 Despite the information gathered through these studies, an analysis of chromatin activation assessed by histone 3 lysine 27 acetylation (H3K27ac), represents a better approach to identify altered regulatory elements and transcription factor (TF) networks in CLL.9-11 In this study, we have generated H3K27ac profiles of stereotyped CLL subsets 1, 2, 4, and 8. In comparison with compared nonstereotyped CLLs, we identified a remarkably distinct profile in stereotyped subset 8, potentially linked to its particularly high risk of Richter transformation (RT), which is the highest among all CLLs.12
Study design
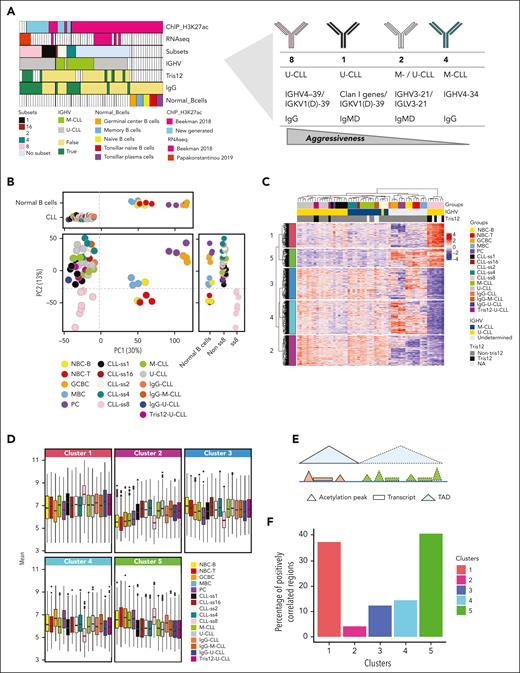
We performed chromatin-immunoprecipitation followed by sequencing (ChIP-seq) with an H3K27ac antibody in sorted B cells from 15 CLL cases from stereotyped subsets 1, 2, 4, and 8. In addition, H3K27ac ChIP-seq profiles from 31 CLLs (including 3 cases of subset 1, 2 cases of subset 4, and 1 case each of subsets 2 and 16) and 15 samples from 5 different normal B-cell subpopulations (n = 3 each) were included from a previous publication from our group.9 Overall, we studied ChIP-seq data from 22 stereotyped CLLs and 24 nonstereotyped CLLs (supplemental Table 1, available on the Blood website). To estimate the functional impact of differential H3K27ac, we integrated our data with RNA-seq data (n = 16) considering the 3D chromatin configuration (Hi-C data from GM12878 and a representative U-CLL case13) using InterTADs tool14 (Figure 1A; supplemental Figure 1; supplemental Table 1; supplemental Methods).
Chromatin activation profiling of stereotyped CLLs. (A) Study design. ChIP-seq and RNA-seq data are labeled by their source (either previously published or generated in this study). (B) Principal component analysis showing components 1 and 2 in CLL subgroups and normal B cells in 2D and 1D plots. Vertical and horizontal dotted lines respectively point to the cut-off value separating CLLs and normal B cells in component 1 and stereotyped subset 8 from the other samples in component 2. (C) Hierarchical clustering is based on the 1185 robust DRs of subset 8 U-CLL cases as compared with other nonsubset U-CLLs. The M-CLL cases and the normal B-cell subpopulations were added to the clustering to aid in the interpretation of H3K27ac profiles. (D) Boxplots showing the mean H3K27ac of the CLL subgroups and normal B-cell subpopulations in each cluster derived from the hierarchical cluster analysis. (E) Scheme of the strategy used to integrate the differential H3K27ac sites with the expression levels within the same TAD. (F) Barplots based on the integrative analysis showing the percentages of positively correlated regions between H3K27ac and gene expression levels in each cluster derived from the hierarchical cluster analysis.
Chromatin activation profiling of stereotyped CLLs. (A) Study design. ChIP-seq and RNA-seq data are labeled by their source (either previously published or generated in this study). (B) Principal component analysis showing components 1 and 2 in CLL subgroups and normal B cells in 2D and 1D plots. Vertical and horizontal dotted lines respectively point to the cut-off value separating CLLs and normal B cells in component 1 and stereotyped subset 8 from the other samples in component 2. (C) Hierarchical clustering is based on the 1185 robust DRs of subset 8 U-CLL cases as compared with other nonsubset U-CLLs. The M-CLL cases and the normal B-cell subpopulations were added to the clustering to aid in the interpretation of H3K27ac profiles. (D) Boxplots showing the mean H3K27ac of the CLL subgroups and normal B-cell subpopulations in each cluster derived from the hierarchical cluster analysis. (E) Scheme of the strategy used to integrate the differential H3K27ac sites with the expression levels within the same TAD. (F) Barplots based on the integrative analysis showing the percentages of positively correlated regions between H3K27ac and gene expression levels in each cluster derived from the hierarchical cluster analysis.
Results and discussion
Initial unsupervised principal component analysis revealed that the main source of the variability (30%) differentiates CLL as a whole from normal B-cell subpopulations (Figure 1B; supplemental Figure 2). Although stereotyped and nonstereotyped CLLs overall clustered together, component 2 of the variability (13%) suggested that subset 8 cases are different from the other CLLs. Next, we performed supervised differential acetylation analyses between each CLL subset vs nonstereotyped CLLs matched by SHM status. Using a stringent false discovery rate (FDR) of 0.0001, we identified 2726 differential regions (DRs) in subset 8 vs U-CLL, 0 DRs in subset 1 vs U-CLL, 3 DRs in 2 vs M-CLL, 0 DRs in 4 vs M-CLL. Out of the 2726 DRs in subset 8, we selected 1185 robust DRs after repeating the test 100 times by applying random resampling (Figure 1C; supplemental Methods; supplemental Figure 3; supplemental Table 2). Overall, both unsupervised and supervised analyses suggest that subsets 1, 2, and 4 show few if any changes compared with IGHV-matched–nonstereotyped CLLs; although we cannot rule out the presence of small-effect changes that cannot be detected owing to the low sample size. However, subset 8 cases are markedly different.
Therefore, we focused the downstream analyses on further characterizing subset 8. Notably, subset 8 cases frequently (∼60%) show trisomy 12,5 which shows differential acetylation profiles in CLL.9,15 To evaluate this relationship, we analyzed the overlap of the subset 8 signature and that established for trisomy 12.9 We detected only 30 DRs in common, which was a nonsignificant overlap (p-hypergeometric test = 0.66), and therefore, the subset 8 signature seemed to be unrelated to trisomy 12.
We subsequently characterized the 1185 DRs and identified 6 subclusters with different patterns, 2 hyperacetylated (clusters 1,5) and 4 hypoacetylated (clusters 2-4) in subset 8. It is worth highlighting that cluster 1 contained regions de novo acetylated in subset 8, whereas the cause of hyperacetylation in cluster 5 seemed to be related to reduced acetylation in nonsubset 8 CLLs as compared with subset 8 (Figure 1C-D). We then designed a strategy to annotate DR to target genes (Figure 1E). First, we linked a given DR to all genes located within a topologically associating domain (TAD) in GM12878, a lymphoblastoid B-cell line widely used to characterize epigenetic patterns in CLL.9 Second, we selected those genes that are differentially expressed between 8 and U-CLL (FDR < 0.01), mining RNA-seq data from partially the same cases.8,9 Finally, we correlated the chromatin acetylation and gene expression levels in the same cases considering only the differentially expressed genes (FDR < 0.01) (Spearman correlation, P value < .05), expecting a positive correlation for target genes. Hyperacetylated regions from clusters 1 and 5 showed high correlation rates with gene expression (78/209 [37.3%] and 63/157 [40.1%]) (Figure 1F). In contrast, hypoacetylated regions showed a lower correlation rate. This analysis suggests that clusters associated with increased H3K27ac in subset 8 frequently contain genes upregulated in this CLL subtype. As cluster 1 was clearly associated with de novo chromatin activation (Figure 1C), we next focused on the 209 regions from this cluster, as they could likely be informative on the specific mechanisms underlying the pathogenesis of subset 8.
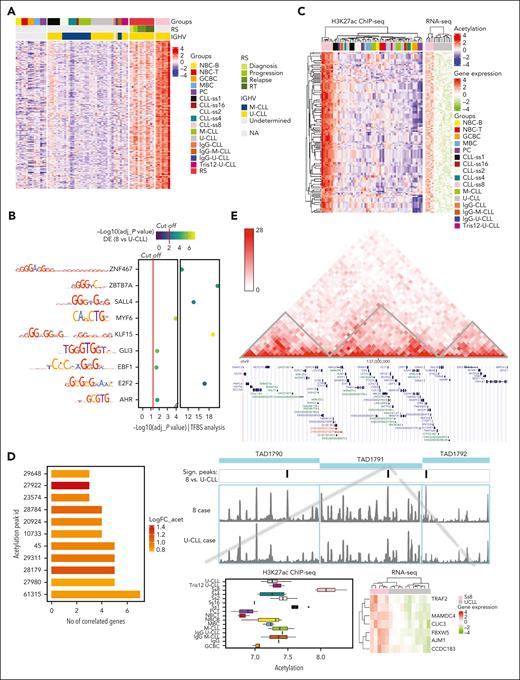
Having seen that subset 8 CLLs have a unique H3K27ac signature, we next wondered whether this signature may be related to their high risk to develop RT. To gain insight into this association, we aggregated recently published longitudinal H3K27ac ChIP-seq data from 4 nonsubset 8 U-CLLs that progressed to RT and included samples at diagnosis, progression, relapse, and RT.11 This comparison revealed a shared pattern between them, both considering the 1185 DRs in subset 8 and the 209 de novo active regions from cluster 1 (Figure 2A; supplemental Figure 3E). These results highlight that the chromatin signature observed in subset 8 cases may potentially contribute to their pronounced tendency to develop RT.
Characterization of the chromatin and transcriptional features of regions de novo acetylated in subset 8 CLLs. (A) Heatmap of the acetylation levels of the 209 de novo active regions including 4 U-CLL cases who experienced Richter transformation with multiple time points (diagnosis, progression, relapse, and Richter transformation). (B) A total of 9 TF binding sites (TFBSs) show statistically significant enrichment (y-axis) in cluster 1 regions and significant correlation with gene expression. The color scale represents the significance of the differences in the expression levels between subset 8 and U-CLL. The left panel shows the binding motifs for each TF. The red lines highlight the cut-off for the statistically significant results in each analysis. (C) Hierarchical clustering is based on 209 regions and associated genes. The heatmap on the left contains the H3K27ac values, whereas that on the right shows the expression values of the associated gene. As some peaks are associated with more than 1 gene, rows of the left heatmap can be duplicated. (D) Barplots showing the H3K27ac peaks that were correlated with the expression levels of more than 2 genes. (E) Hi-C map for 1 representative nonstereotyped U-CLL case and genome browser display of TAD1791 containing peak-61315 (including also the flanking TADs 1790 and TAD1792 as controls). We show the H3K27ac coverage plot of a representative subset 8 and nonstereotyped U-CLL. On the left, a zoom-in panel of the significant peak-61315 within TAD1791 including a boxplot of the H3K27ac levels and a heatmap with the gene expression levels of the 7 associated genes.
Characterization of the chromatin and transcriptional features of regions de novo acetylated in subset 8 CLLs. (A) Heatmap of the acetylation levels of the 209 de novo active regions including 4 U-CLL cases who experienced Richter transformation with multiple time points (diagnosis, progression, relapse, and Richter transformation). (B) A total of 9 TF binding sites (TFBSs) show statistically significant enrichment (y-axis) in cluster 1 regions and significant correlation with gene expression. The color scale represents the significance of the differences in the expression levels between subset 8 and U-CLL. The left panel shows the binding motifs for each TF. The red lines highlight the cut-off for the statistically significant results in each analysis. (C) Hierarchical clustering is based on 209 regions and associated genes. The heatmap on the left contains the H3K27ac values, whereas that on the right shows the expression values of the associated gene. As some peaks are associated with more than 1 gene, rows of the left heatmap can be duplicated. (D) Barplots showing the H3K27ac peaks that were correlated with the expression levels of more than 2 genes. (E) Hi-C map for 1 representative nonstereotyped U-CLL case and genome browser display of TAD1791 containing peak-61315 (including also the flanking TADs 1790 and TAD1792 as controls). We show the H3K27ac coverage plot of a representative subset 8 and nonstereotyped U-CLL. On the left, a zoom-in panel of the significant peak-61315 within TAD1791 including a boxplot of the H3K27ac levels and a heatmap with the gene expression levels of the 7 associated genes.
As chromatin activation patterns are frequently associated with binding sites of particular TF,9,10 we next performed a TF binding site analysis. The regions from cluster 1 were significantly enriched (adj-P value < .05) in binding sites of ZNF467, ZBTB7A, SALL4, MYF6, KLF15, GLI3, EBF1, E2F2, and AHR (Figure 2B) whose expression was upregulated in subset 8 in comparison with U-CLL. Most of them have previously been associated with CLL, such as MYF6 which is overexpressed in trisomy 12 cases,16 or other hematologic malignancies, such as ZBTB7A, which is mutated in acute myeloid leukemia.17 This analysis suggests that a few overexpressed TFs may be responsible for the de novo chromatin activation pattern observed in subset 8.
We next focused on the downstream effects of chromatin activation in subset 8 by characterizing the potential target genes. Of the 209 regions of cluster 1, 78 unique peaks were significantly correlated with the expression of 113 unique genes, which also were overexpressed in subset 8 (Figure 2C; supplemental Table 3). Among the top 5 differentially expressed genes (ie, HLA-DRA, ZDHHC19, LINGO3, NDUFA4L2, and GZMM), mitochondrial protein NDUFA4L2 regulates cell survival associated with poor prognosis in human cancers.18 Moreover, ZDHHC19 mediates palmitoylation and provides signals on STAT3 regulation,19 which plays a critical role in CLL cell survival.20
Beyond individual gene functions, we observed that 11 out of 78 activated regions were correlated with more than 2 genes, suggesting that several genes within a TAD could be associated with subset 8 pathogenesis through local coupregulation (Figure 2D). Among them, the acetylation peak 61315 located in chromosome 9 showed a positive correlation with the expression of 7 transcripts (6 coding genes CLIC3, FBXW5, AJM1, MAMDC4, CCDC183, and TRAF2 as well as 1 unannotated transcript) within its host TAD (Figure 2D-E). In this case, although peak 61315 was the only significantly different one, we observed 7 additional peaks in the region with increased H3K27ac in subset 8 (supplemental Figure 4), suggesting that coupregulated genes in this area are not only affected by 1 peak but by the activation of several regulatory elements within the TAD. As controls, we used the TADs next to TAD1791 (TAD1790 and TAD792), which showed 1 differential H3K27ac peak each that was not correlated with the expression of the genes within their host TAD or the adjacent TAD1791. To examine the overexpressed genes in subset 8 regulated by peak-61315, TRAF2 was a remarkable example because this gene suppresses death-receptor–mediated apoptosis and cooperates with BCL2 promoting CLL/small lymphocytic lymphoma in mice.21 Further supporting the role of TRAF family, TRAF6, TRAF4, and TRAF1 have been described to be overexpressed in U-CLL.22 Additionally, other coupregulated genes within peak 61315, such as CLIC3, FBXW5, and CCDC183 have been associated with poor prognosis in human cancers.23-25
Taken together, our systematic ChIP-seq analysis in stereotyped and nonstereotyped CLLs reveals that subset 8 cases are characterized by a distinct chromatin activation signature. This finding further supports the distinct biological nature of subset 8 and may help to understand the particularly aggressive clinical behavior of this peculiar CLL variant, including its predisposition to develop RT.
Acknowledgments
This work was partially developed at the Centro Esther Koplowitz (Barcelona, Spain).
This research was funded by the European Research Council under the European Union’s Horizon 2020 research and innovation program (Project BCLLATLAS, grant agreement 810287), Fundació la Marató de TV3, Generalitat de Catalunya Suport Grups de Recerca AGAUR 2017-SGR-736, CIBERONC (CB16/12/00225), the Accelerator award CRUK/AIRC/AECC joint funder-partnership, the Hellenic Precision Medicine Network in Oncology, and the project ODYSSEAS (Intelligent and Automated Systems for enabling the Design, Simulation, and Development of Integrated Processes and Products) implemented under the “Action for the Strategic Development on the Research and Technological Sector,” funded by the Operational Programme “Competitiveness, Entrepreneurship and Innovation” (NSRF 2014-2020) and cofinanced by Greece and the European Union.
Authorship
Contribution: M.T. designed the research, analyzed the data, performed the study, and wrote the manuscript; V.C., B.G.-T., N. Pechlivanis, and F.N. provided data and assisted in data interpretation; N.R. performed the ChIP-seq libraries; N. Papakonstantinou, A.C., F.P., and E.C., assisted in research, data analysis, and interpretation; N.S. provided patient samples; and J.I.M.-S. and K.S. designed, supervised the research, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: José Ignacio Martín-Subero, Institut d'Investigacions Biomèdiques August Pi i Sunyer, Universitat de Barcelona, Rossello 153, 08036 Barcelona, Spain; e-mail: imartins@recerca.clinic.cat; and Maria Tsagiopoulou, Centro Nacional de Análisis Genómico, Carrer de Baldiri Reixac, 4, torre i, 08028 Barcelona, Spain; e-mail: maria.tsagiopoulou@cnag.crg.eu.
References
Author notes
∗J.I.M.-S. and K.S. contributed equally to this study.
All the generated raw sequencing data (fastq files) are available through EGA under the accession number EGAS00001006457. All the scripts for the analysis of the raw data until the visualizations are available on Github (https://github.com/MariaTsayo/H3K27ac_ss8) with a detailed description of each script. Finally, the matrixes that were used in this study as well as the metadata are available on Zenodo under the https://doi.org/10.5281/zenodo.6865838. Regarding the integration with gene expression, 11 RNA-seq data samples from U-CLL cases were available at ICGC and the 5 subset 8 cases were available at: https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6962/. Hi-C data from our recent study were visualized through a 3D genome browser (http://3dgenome.fsm.northwestern.edu/) for 1 representative U-CLL case3 as well as from GM128781 and were used as the topologically associated domains.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal