Abstract
Thrombotic microangiopathy (TMA) encompasses various genetically-driven diseases. The emergence of ultrafast genomic sequencing has recently opened up new avenues of research for genetic investigations in the setting of intensive care units. TMA is likely to be a suitable focus for fast-track genomic sequencing. By establishing an expeditious molecular diagnosis of patients with the complement-dependent hemolytic uremic syndrome, fast-track genomic sequencing allows for the timely implementation or withdrawal of anti-C5 treatment while averting unnecessary, costly, and potentially harmful therapy in patients testing negative for the syndrome. Furthermore, genomics has the potential to reshape the taxonomic classification of TMA owing to comprehensive genomic analysis. The most significant results from such analysis can be categorized as (1) new descriptions of genetic diseases previously not recognized as associated with TMA and (2) an enrichment of the phenotypic spectrum of diseases traditionally related to TMA. The latter draws on the concept of retrophenotyping, wherein genomic investigation precedes full clinical description. By taking precedence over a phenotypic approach, an unbiased genomic-focused analysis maximizes the chances of discovering new descriptions of a given variant. Presented here are 4 cases of TMA which highlight these issues and substantiate the promise of fast-track genomic sequencing.
Introduction
Thrombotic microangiopathy (TMA) represents a great clinical conundrum because it encompasses a myriad of causes ranging from Shiga toxin–mediated TMA to a wide spectrum of genetically-driven diseases. Furthermore, clinicians face the additional challenge of confidently discriminating between different etiologies based solely on routine clinical evidence, often in the face of urgently needed specific therapy.
Over the last few years, next-generation sequencing (in particular exome sequencing [ES] or genome sequencing [GS]) has steadily established itself as an essential step in the diagnostic workup of patients in numerous settings.1-5 The turnaround time, defined by the length of time between sample handling and the genetic sequencing report, has previously hindered the diffusion and implementation of genomic explorations in the adult acute care setting significantly. Until recently, this delay was counted in months, which is clearly at odds with the requirements of acute care management.6
The emergence of ultrafast GS in neonatal intensive care units should, therefore, be welcomed as a new milestone.7-11
Ultrafast genomic testing is also bound to find applications in adult acute care. One such application includes patients with TMA who may benefit from extensive and expeditious genomic sequencing. Indeed, once thrombotic thrombocytopenic purpura (TTP) and Shiga toxin–mediated hemolytic uremic syndrome (HUS) have been ruled out, one of the key issues is to identify complement-mediated TMA (CM-TMA) requiring anti-C5 therapy. Hence, for TMA with acute kidney injury, especially in the context of malignant hypertension, clinicians face a dilemma, which is whether to withhold anti-C5 therapy and run the risk of patients suffering irreversible renal damage or to initiate unwarranted anti-C5 therapy and needlessly expose patients to the increased likelihood of encapsulated pathogen-related infections; not to mention the costs involved. From this standpoint, ultrafast GS may help determine whether TMA is related to a Mendelian disease and further delineate between variants affecting genes involved in complement regulation (eg, complement factor H (CFH), CFI, C3, CFB, MCP, and CFHRs) and other noncomplement genetic disorders (eg, MMACHC, DGKE, and INF2), a key step in selecting suitable candidates for anti-C5 therapy.12-14 The clinical strategy of “reverse phenotyping” has also the potential to reshape the nosological categorization of TMA by (1) ascertaining molecular diagnosis, (2) describing genetic diseases previously not recognized as associated with TMA,15 and (3) enriching the phenotypic description of diseases traditionally known to be complicated with TMA.
Here, we describe 4 cases of TMA that highlight how incorporating fast genomic explorations may transform the diagnostic workup and therapeutic approach for patients with TMA and discuss some of the common practical issues and dilemmas which may arise when implementing such strategies. They provide a glimpse of how “fast-track genomic” investigations might transform our understanding and management of TMA.
Illustration of cases
Supplemental Table 1, available on the Blood website, summarizes the phenotypes at presentation and genetic variations between these 4 cases. A written and informed conscent was obtained for all patients.
Case study 1
In 2018, a 26-year-old male patient with a 4-year history of high blood pressure (BP) was admitted for malignant hypertension (210/140 mm Hg), complicated by kidney dysfunction (plasma creatinine [PCr] =139 μmol/L, estimated glomerular filtration rate [eGFR] = 60 mL/min per 1.73 m2) and biological TMA. A kidney biopsy found mixed arteriolar and glomerular TMA, along with mild arteriolosclerosis and moderate interstitial fibrosis. TTP, Shiga toxin–producing Escherichia coli (STEC-HUS), and secondary HUS were ruled out. Given that renal function was initially stable, nephroprotective therapy alone was initiated. The patient’s kidney function deteriorated gradually over the next 2 years, with an eGFR of 41 mL/min per 1.73 m2 prompting ES. Three weeks later, ES reported a heterozygous compound MMACHC/PRDX1 variation (PRDX1 NM_002574.3: c.515-1G>T and MMACHC NM_015506.3: c.566G>A). The PRDX1 mutant allele causes an MMACHC secondary epimutation in patients with cobalamin C (cblC) deficiency.16 Because the position of this variant had never been previously described, additional functional tests were ordered to ascribe the phenotype to the documented variants. They were consistent with the diagnosis of cblC, demonstrating hyperhomocysteinemia (346 μmol/L, N < 15), elevated plasmatic and urinary methylmalonic acid, and low methionine. The MMACHC/PRDX1 variations could thus be ranked American College of Medical Genetics and Genomics class 4. The patient received a diagnosis of methylmalonic aciduria with cblC.
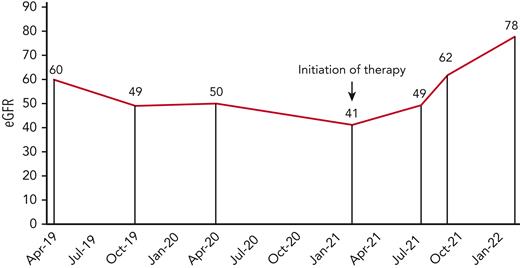
Kidney function (eGFR) in a patient with MMACHC-related disease before and after hydroxocobalamin therapy.
Kidney function (eGFR) in a patient with MMACHC-related disease before and after hydroxocobalamin therapy.
Impact of the genetic diagnosis
Modification in therapeutics and clinical improvement: the patient was started on hydroxocobalamin and betaine supplementation 4 weeks after the initial consultation. His BP improved and there was an improvement in his kidney function from 41 to 78 mL/min per 1.73 m2 over a 9-month follow-up (Figure 1).
Family counseling: the patient’s elder sibling had previously been diagnosed with high BP at the age of 25 years and had received a diagnosis of vascular chronic kidney disease (CKD) on follow-up. TMA had not been reported. The patient’s 2 younger siblings displayed normal BP and kidney function. Targeted panel sequencing (TPS) in the oldest sibling demonstrated the same heterozygous compound MMACHC/PRDX1 variation.
Retrophenotyping: brain magnetic resonance imaging (MRI) displayed widespread cortical atrophy predominating on the left side, the cerebellum hemispheres, vermis, and corpus callosum. In addition, the corpus callosum enclosed lacunar-like lesions. There was no evidence of cardiac or ophthalmological involvement related to MMACHC.
Case study 2
A 26-year-old White man was hospitalized in 2017. He had no relevant personal medical history. He presented headaches revealing high BP (200/140 mm Hg). His clinical examination was found unremarkable apart from patchy alopecia and absent eyebrows. Laboratory tests revealed a normal blood count with no sign of TMA. The patient underwent a kidney biopsy to explore glomerular proteinuria of 2 g/g without hematuria or kidney dysfunction (PCr = 84 μmol/L, eGFR = 110 mL/min per 1.73 m2). The kidney biopsy demonstrated glomerular and arteriolar TMA. TTP, STEC-HUS, and other forms of TMA were ruled out.
Homocysteine plasma levels were moderately elevated (41 μmol/L, N < 15). An initial TPS exploring an alternative pathway gene (including MMACHC) was found to be negative. Subsequently, an ES reported a heterozygous pathogenic variant in SOX18 (NM_018419.2:c.474_476delinsAG, p.Lys159Glyfs∗19) leading to the clinical diagnosis of hypotrichosis-lymphedema-telangiectasia-renal defect syndrome (Online Mendelian Inheritance in Man #137940).
Impact of the genetic diagnosis:
Retrophenotyping: hypotrichosis-lymphedema-telangiectasia syndrome (HTLS) is a rare hereditary disorder with recessive or dominant inheritance and with significant phenotypic variability. Other characteristic features of HTLS are defects in hair follicle development (100%) resulting in scalp hair loss, absence of eyebrows or eyelashes, lymphedema in the lower limbs and eyelids (90%), and cutaneous telangiectasias (77%). Other findings, such as bilateral hydroceles in men and dysmorphic facial features,17 are less commonly observed. After extensive retrophenotyping, the patient was found to have no other features consistent with HTLS, except for generalized hypotrichosis characterized by sparse hair on the vertex and temples (Figure 2), almost no eyebrows, and very short eyelashes.
Identification of a new autosomal dominant variant of SOX 18: in 2003, Irrthum et al identified SOX18 as the gene underlying both recessive and dominant HTLS.18SOX18 is located on chromosome 20 (20q13.33), where it encodes a transcription factor involved in the development of the vasculature and lymphatic system. As per our observation, the frameshift variant resulted in a premature stop codon at amino acid 178.
Enrichment of the repertoire of TMA: to date, only 2 patients with a pathogenic variant affecting SOX18 have exhibited kidney disease out of a total of 10 unrelated patients reported in the literature. In both cases, kidney disease was detected during childhood and renal histological features were consistent with TMA. Of note, the 2 patients developed end-stage kidney disease.19
Family counseling: Sanger analysis in the patient’s parents demonstrated that the SOX 18 variant was inherited by his mother who also presented with hypotrichosis but without the evidence of kidney disease.
Informing the therapeutic strategy: the diagnosis of SOX18-related TMA may limit costly and unnecessary therapeutic treatments, most notably eculizumab, even though TMA was evidenced on renal biopsy. Of note, whole ES–guided retrophenotyping helped to identify and exclude potentially life-threatening vascular complications in HTLS, namely pulmonary hypertension and aortic dilatation.17,20
The pathophysiological pathway connecting SOX18 and TMA is still an area of research; however, SOX18 is a well-recognized transcription factor involved in endothelial cell differentiation.
Case study 3
A 22-year-old female patient was hospitalized for TMA in the context of malignant hypertension and mild kidney dysfunction (PCr = 107 μmol/L, eGFR 60 mL/min per 1.73 m2). She had a known 5-year personal history of hypertension. Her eldest sister had a remarkable history of hypertension and CKD, presumed to be hypertensive nephrosclerosis solely based on clinical criteria. Her 4 remaining siblings had no history of high BP or CKD.
Plasma exchange was discontinued once ADAMTS13 activity was found to be normal. Kidney biopsy demonstrated a single lesion of focal and segmental glomerulosclerosis, severe vascular lesions, arteriolar TMA, and 50% interstitial fibrosis. Functional exploration of the complement alternative pathway was unremarkable. Secondary hypertension was ruled out. No anti-C5 therapy was initiated as TMA resolved once hypertension was controlled and kidney function remained stable.
She was lost to follow-up and was readmitted 2 years later, when 12 weeks pregnant, for malignant hypertension, including severe hypertensive retinopathy, biological TMA, and end-stage CKD. An abortion on medical grounds was agreed and permanent hemodialysis was initiated.
Four years later, one of her sisters was referred to our department for kidney dysfunction and biological TMA. ES revealed a homozygous pathogenic variant in TTC21B (NM_024753.4:c.626C>T, p.Pro209Leu). The homozygous variation was confirmed via Sanger sequencing for the proband patient. Both the proband patient and her sibling received a final diagnosis of type 12 nephronophthisis.
Impact of the genetic diagnosis:
Family counseling with 3 of the 6 siblings affected: because of its recessive heredity, we could reassure the patients regarding the risk of their children getting affected.
Identification of a new phenocopy of nephronophthisis: at our center, we identified 7 cases of nephronophthisis misclassified as hypertensive kidney disease.15 Three patients presented initially with the hallmark of biological TMA whereas histology yielded arteriolar TMA on the renal biopsy in 3 other cases. The phenotype of these patients included hardly any of the characteristics of nephronophthisis consistent with its canonical description. Nephronophthisis was unlikely to have ever been diagnosed without broad genomic investigations.
Case study 4
A 53-year-old male patient with no relevant medical history received a consultation in the emergency room for malignant hypertension (200/120 mm Hg), kidney dysfunction (PCr = 680 μmol/L, blood urea nitrogen = 40 mmol/L), and biological TMA. ADAMTS13 activity was normal. STEC-HUS was excluded.
Despite 19 rounds of plasma exchanges and steroids, the patient could not be weaned from hemodialysis and evidence for TMA persisted. On kidney biopsy, half of the glomeruli were found to be sclerotic, and other glomeruli demonstrated TMA-related lesions. Arteriolar TMA was also reported. Eculizumab was initiated 4 weeks after the initial presentation. The patient was referred to our center for genetic exploration. Within 3 weeks, ES results yielded a pathogenic, that is, American College of Medical Genetics and Genomics class 5 variant, affecting CFH (NM_000186.4:c.3047A>G).
Impact of the genetic diagnosis:
Family counseling: no family history of TMA was reported. Exploratory-targeted genetic testing was undertaken on the patient’s 2 daughters who were of child-bearing age. The youngest daughter was found to be carrying the CFH variation even though she had a history unremarkable for TMA, hypertension, or kidney disease. She was informed of the implications of bearing this variant, including the incomplete predictability of developing TMA because of its variable penetrance.
Informing the therapeutic strategy: ES confirmed the indication of anti-C5 therapy within 21 days of genetic investigations. Unfortunately, the treatment was introduced at a late stage so chronic renal damage was already present, and the patients remained on permanent hemodialysis. Nonetheless, the genetic diagnosis confirmed the indication of eculizumab in the event of kidney transplantation.
Discussion
These cases reinforce the transformative benefit of prompt comprehensive genetic testing in the setting of TMA. Despite extensive investigations consistent with current guidelines, in each case, the final diagnosis could only be ascertained by ES. In our case studies, the turnaround time from sample receipt to ES report ranged from 3 to 8 weeks. Most of the cases solved after ES could be categorized as cold cases, with patients affected by long-standing renal disease. However, the data for these cases were collected to examine the potential of fast genomics applied to TMA and to serve as a basis for its standardized and timely implementation for future cases.
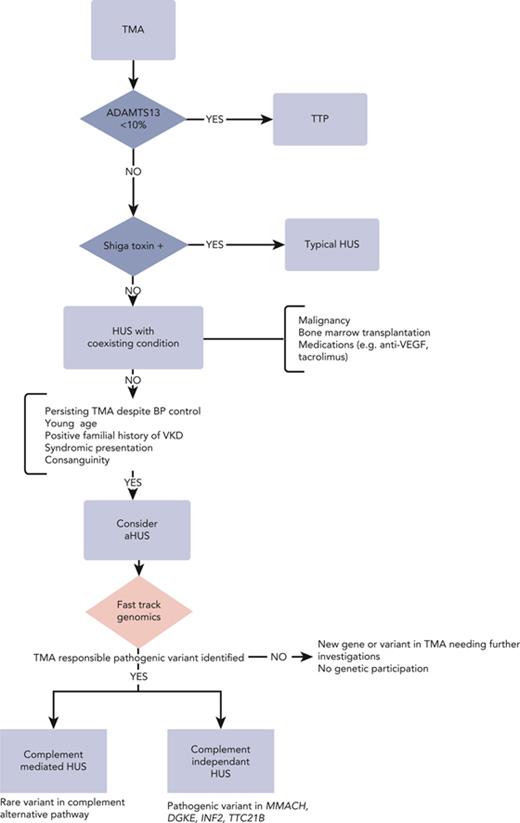
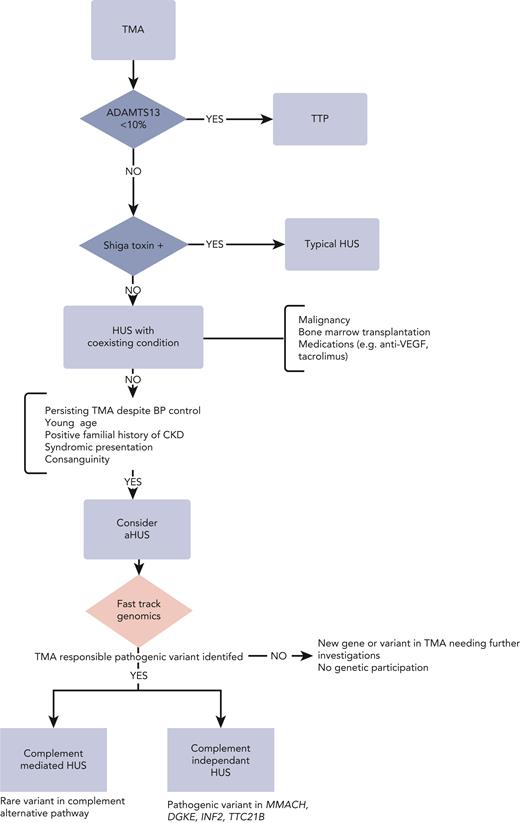
Initial diagnostic workup when faced with TMA: rule out TTP
TMA comprises a myriad of clinical entities with distinct pathophysiological mechanisms sharing microangiopathic mechanical hemolytic anemia and thrombocytopenia as common traits. Given its distinctive management and dismal prognosis in case of delayed identification, during the initial diagnostic workup, emphasis is placed on delineating TTP from other causes of TMA. The definitive diagnosis of TTP is ascertained by demonstrating greatly reduced (<10%) levels of ADAMTS13. Results of ADAMTS13 activity level pending, PLASMIC or French scores, based on readily accessible clinical and biological variables, have been devised to assist the clinician in predicting the likelihood of TTP. The prevailing paradigm is to promptly initiate plasma exchanges whenever the diagnosis is suspected. The mechanism underpinning TTP is predominantly acquired and autoimmune (95% of the cases) owing to circulating autoantibodies directed against ADAMTS13. Yet, in selected populations, namely pediatric and pregnant patients, congenital TTP (cTTP; Upshaw-Schulman syndrome) resulting from recessive biallelic ADAMTS13 pathogenic variants may account for up to 25% to 50% of the cases.21 Although plasma exchanges are the cornerstone of both cTTP and immune TTP (iTTP),22-24 the latter form of TTP differs in that it requires immunosuppressive therapy based on steroids and rituximab to mitigate the risk of relapse,25 as well as anti–von Willebrand therapy based on caplacizumab for its temporizing effects.23,26,27 In contrast, ADAMTS13 replenishment suffices in cTTP.28 However, discriminating between both forms of TTP relies heavily on documenting an ADAMTS13 inhibitor whose identification may be obscured in severe hemolysis. Furthermore, the absence of an ADAMTS13 inhibitor does not preclude the diagnosis of iTTP, and ultimately the diagnosis of cTTP rests on the kinetics of ADAMTS13 activity level, which typically remains very low after the flare. In specific settings,29,30 clinicians may value genetic testing to distinguish cTTP from iTTP to avoid superfluous treatments and the risks involved, such as bleeding complications with caplacizumab and infections with immunosuppressive therapies.
Finally, in a prospective study on eculizumab discontinuation in patients with atypical HUS (aHUS), the only patient with no germ line complement mutation who relapsed was shown to have inherited ADAMTS13 deficiency.31
Once TTP and Shiga toxin–mediated HUS have been excluded, the next step is to identify CM-TMA.
Identification of CM-TMA: a clinical challenge
Typically, CM-TMA, previously termed aHUS, is established by the presence of TMA and acute kidney injury after the exclusion of all other conditions associated with secondary TMA.31 One of the causes of secondary TMA is malignant hypertension, but conversely, malignant hypertension is highly prevalent in patients with CM-TMA, ranging from 34% to 55% of cases, irrespective of its cause.32,33 Even though studies suggest that subtle clinical or biological cues, such as diastolic BP or platelet counts,34 could imply the existence of CM-TMA, no reliable score can confidently discriminate CM-TMA from other causes of TMA.
Testing for inhibitory antibodies to complement regulatory proteins, mainly antibodies to CFH, is mandatory in the context of suspicion of CM-TMA. Yet, anti–factor H antibodies account for <6% of adult CM-TMA, suggesting a very wide scope for genetic causes of TMA.30,35,36 Broad serum complement assessments (C3 and C4 levels, and total complement activity) are known for their low sensitivity and specificity.30,35,36 In 40% of the cases, C3 levels are within the normal range at presentation.36 Histopathological investigations have failed to identify cause-specific features, or they have produced conflicting results.34,37 Ex vivo testing of C5b-9 formations on microvascular endothelial cells to detect patients with CM-TMA is promising, but it is still in the early stages of development.34,38,39 Likewise, hyperhomocysteinemia is recognized as a hallmark of cblC. Some patients, however, only exhibit slightly elevated levels, and renal failure is associated with increased homocysteine levels.40 Ultimately, biomarkers,despite their usefulness, are not a substitute for genetic testing.41
Use of genomics to guide clinical management and therapeutics in TMA
By ruling out the diagnosis of CM-TMA, ES/GS spares the patient unnecessary and potentially harmful anti-C5 therapy and can allow for the swift implementation of specific therapy in certain cases. In the case studies, genetic testing finally ended years of misguided clinical management by producing molecular evidence. In the setting of MMACHC-related disease, swift initiation of hydroxocobalamin is crucial, permitting significant improvement of renal injury (case 1).42,43 Therapeutic indications may extend beyond mere kidney-related outcomes, and hydroxocobalamin therapy remains warranted even if patients progress to end-stage CKD. In addition to TMA and CKD, cblC can cause neurological symptoms, retinal degeneration, and thromboembolic and cardiopulmonary events.42,44,45 Genetic testing can also substantiate the indication for anti-C5 therapy, both for immediate use and future prevention of CM-HUS relapse in the setting of kidney transplantation (case 4). It can also offer a unifying interpretation of seemingly distinct clinical manifestations. The patient in case study 3 underwent multiple investigations, including a liver MRI and liver biopsy, to explore chronic cholestasis, a typical feature of nephronophthisis.
Comprehensive genetic explorations to enable family counseling and prevention
Genome investigations allowed for family counseling, testing, and eventually selection for prospective living-related kidney transplantation. For instance, in case study 1, one of the proband patient’s older brothers had been diagnosed with hypertension in his 20s and was followed up for CKD stage 3, presumed to be nephrosclerosis.Focused genetic testing targeting MMACHC/PRDX1 genes ascertained the diagnosis of cblC. He was immediately started on hydroxocobalamin therapy. Thanks to family screening in case study 3, 3 sisters out of 6 siblings were diagnosed with nephronophthisis type 12. In case study 4, the 2 adult daughters of the proband patient were screened for a CFH pathogenic variation.
Genomics explorations to expand medical knowledge
An additional benefit of extended genetic testing is that it may potentially refine the phenotypic description of a disease related to a given gene. Patients with cblC typically display a wide array of neurologic-, hematologic-, and heart-related features, along with TMA, with a predominantly childhood onset.44,46 In contrast, extrarenal manifestations were absent in case study 1. Compared with the previous case series, the patient may appear to be simply an outlier. Alternatively, it questions whether cblC may be underdiagnosed, going unrecognized and/or masquerading as CM-HUS. At the least, this case, together with recent reports, establishes cblC as a plausible candidate whenever first-line explorations for causes of TMA are inconclusive in adult populations, with potentially major therapeutic and prognostic consequences, even if characteristic extrarenal manifestations are absent.47 Cases 2 and 3 also epitomize how ES can uncover new genes previously unrelated to a given phenotype. Neither HTLS nor nephronophthisis were hitherto known to harbor TMA within their clinical spectrum, thereby expanding their phenotypic description. In case 3, this heralded the emergence of a new vascular phenotype, “nephroangionophthisis.”15 More broadly, these cases point to the phenomenon of phenocopies or locus heterogeneity,15,48,49 the terms coined to define diseases that are indiscernible from a phenotypic perspective but can be ultimately traced to distinct genotypes.50
Limitations of pangenomic explorations
The expense of GS/ES is often cited as a major limitation for its widespread implementation, but costs have recently plummeted.51,52 To date, in France, the price charged for an ES is €1100 (US $1294), comparable to that of a single brain MRI in the United States.53 In the context of TMA, opting for anti-C5 therapy also carries a considerable financial burden and ES/GS costs may be offset by the savings entailed.54,55 For this reason and because GS/ES requires specific bioinformatic resources, some authors favor TPS, which also boasts greater sequencing depth and accuracy. Clinicians should also be wary of ethical concerns which may arise from the misuse of ES/GS data leading to discriminatory practices, namely in terms of health insurance and employment.
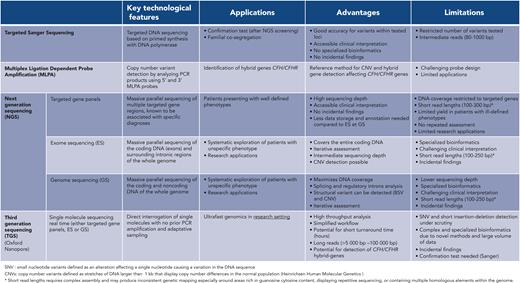
ES/GS carry challenges of their own (Figure 3). They may give rise to variant of uncertain significance,56 whose handling is notoriously complex. On occasion, the variant of uncertain significance can be reclassified by additional cosegregation studies which involve phenotyping and genotyping relatives. Such investigations are time- consuming, may delay in reporting the result, and can prove to be inconclusive. Neither retrophenotyping nor cosegregation is infallible. Retrophenotyping can be undermined in the case of variable disease expressivity, that is, a differing phenotype between 2 individuals with a common phenotype, whereas cosegregation can prove inconclusive in case of variable penetrance. In this setting, functional data can intervene to provide decisive data. Genotyping cannot supplant a comprehensive clinical exploration, it is the interplay of genotyping with (retro)phenotyping that is most likely to provide compelling results (case 1). In aHUS cohorts, ∼40% of patients considered to be suffering with CM-TMA had neither a rare complement gene variant nor anti–factor H antibodies based on targeted gene panels.31,33,57 One can surmise that an unspecified proportion of these patients with aHUS analysis may have a genetic cause unrelated to complement proteins, which can only be revealed through pangenomic explorations. Alternatively,a subset of these patients might not have a form of aHUS caused by a disease of Mendelian inheritance. Nevertheless, negative genetic explorations concerning the complement remain a crucial route for patient management because they predict a low risk of relapse after eculizumab withdrawal and kidney transplantation. Current ES/GS cannot reliably identify complex hybrid rearrangement affecting CFHR1 or CFHR3 and CFH genes,58-60 yet they account for <5% of CM-TMA.36,61 To overcome this limitation, a multiplex ligation-dependent–probe amplification (reference method), needs to be considered (Figure 3). Finally, clinicians should be cognizant of other general limitations inherent to ES/GS, such as the short read lengths produced lead to inconsistent genetic mapping especially around areas rich in guanosine cytosine content, displaying repetitive sequencing, or containing multiple homologous elements within the genome. So-called third-generation sequencing has driven the emergence of sequencing based on long reads (>10 kilobase pairs) to circumvent these constraints.
Comparative overview of sequencing technologies. Small nucleotide variants (SNVs) defined as an alteration affecting a single nucleotide causing a variation in the DNA sequence copy number variants (CNVs) defined as stretches of DNA larger than 1 kilobase that displays copy number differences in the normal population (Heinrichsen Human Molecular Genetics). ∗Short read lengths require complex assembly and may produce inconsistent genetic mapping, especially around areas rich in guanosine cytosine content, displaying repetitive sequencing, or containing multiple homologous elements within the genome.
Comparative overview of sequencing technologies. Small nucleotide variants (SNVs) defined as an alteration affecting a single nucleotide causing a variation in the DNA sequence copy number variants (CNVs) defined as stretches of DNA larger than 1 kilobase that displays copy number differences in the normal population (Heinrichsen Human Molecular Genetics). ∗Short read lengths require complex assembly and may produce inconsistent genetic mapping, especially around areas rich in guanosine cytosine content, displaying repetitive sequencing, or containing multiple homologous elements within the genome.
The issue regarding the indication of genetic investigations and how they should be articulated with other tests can only be settled using a first-stage strategy and available technology, one that combines (1) ES/GS sequencing that can identify genes unsuspected beforehand, and (2) reverse phenotyping that can uncover subtle, hitherto unnoticed, clinical signs based on molecular findings. This approach is best exemplified by case study 1, in which the genetic demonstration of cblC disease helped unravel neurological complications, which in turn further substantiated the diagnosis. We propose a working algorithm (Figure 3) which incorporates genomic testing. Clinicians should be wary of traditional predictors of positive genetic investigations such as young age and positive family history. Case study 4 was of a 53-year-old male patient with a diagnosis of CM-TMA, whereas familial anamnesis was initially negative in case studies 1, 2, and 4. Accordingly, investigators recently reported that 25% of patients with proven genetic kidney disease have a negative family history62 and this can reach 80% in TMA.36 In routine clinical practice, its timely implementation may offer key evidence for otherwise tentative clinical diagnoses with consequential ramifications in terms of therapeutic management, prognosis, and family counseling. In research-oriented protocols, next-generation sequencing is bound to uncover new genes and mechanisms of disease and expand the phenotypic inventory of known conditions. Finely tuned models are needed to pinpoint the appropriate timing and indication of these explorations before ES/GS is widely adopted as a first-tier diagnostic framework. Although financial modeling is currently lacking, both the sharp drop in costs and the potential savings entailed are gradually tipping the balance in favor of ES/GS before moving forward to long read–based technologies. The current ES turnaround time is gradually declining and given recent advances, a 3-day delay seems an achievable sweet spot. Although this compares unfavorably with the time-to-result of neonatal intensive care units,11 it might be appropriate for the conditions described here, enabling clinicians to implement adequate treatments beforehand and opt out of unwarranted therapies. Furthermore, the turnaround time is bound to reduce dramatically with the arrival of third-generation sequencing very soon (Figure 3). A preliminary phase of extensive genomic testing may prove necessary to delineate its relevant applications and clinical profiles.
Acknowledgments
The authors thank Laure Raymond, Marine Dancer, Alexandre Cez, and Jean-François Benoist for the critical appraisal and review of the manuscript and expert commentary on genetic technological aspects (Laure Raymond and Marine Dancer) and MMACHC-related disease (Jean-François Benoist).
Authorship
Contribution: A.D., L.M., and C.R. were responsible for data curation; and all the authors were responsible for manuscript drafting, revision, and critical appraisal.
Conflict-of-interest disclosure: C.R. receives lecture fees from Alexion Pharma France and travel grants. L.M. receives lecture fees from Travere Pharmaceutical and Sanofi Pharma and travel grant from Sanofi France Pharma. E.R. receives research grants and personal fees for expertise from Alexion Pharma France and research grants from Roche. A.D. declares no competing financial interests.
Correspondence: Alice Doreille, Service de Soins Intensifs et Rein Aigu, Hôpital Tenon, Assistance Publique–Hôpitaux de Paris, 75020 Paris, France; e-mail: alice.doreille@aphp.fr; and Laurent Mesnard, Service de Soins Intensifs et Rein Aigu, Hôpital Tenon, Assistance Publique–Hôpitaux de Paris, 75020 Paris, France; e-mail: laurent.mesnard@aphp.fr.
References
Author notes
The online version of this article contains a data supplement.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal