Key Points
Intrathymic delivery of AAV results in vector integration within TCR genes at RAG-induced DNA breaks produced during V(D)J recombination.
This “targeting” approach opens therapeutic avenues for long-term AAV gene transfer in dividing cells without any toxic conditioning.
Abstract
Adeno-associated virus (AAV) vectors have been successfully exploited in gene therapy applications for the treatment of several genetic disorders. AAV is considered an episomal vector, but it has been shown to integrate within the host cell genome after the generation of double-strand DNA breaks or nicks. Although AAV integration raises some safety concerns, it can also provide therapeutic benefit; the direct intrathymic injection of an AAV harboring a therapeutic transgene results in integration in T-cell progenitors and long-term T-cell immunity. To assess the mechanisms of AAV integration, we retrieved and analyzed hundreds of AAV integration sites from lymph node-derived mature T cells and compared these with liver and brain tissue from treated mice. Notably, we found that although AAV integrations in the liver and brain were distributed across the entire mouse genome, >90% of the integrations in T cells were clustered within the T-cell receptor α, β, and γ genes. More precisely, the insertion mapped to DNA breaks created by the enzymatic activity of recombination activating genes (RAGs) during variable, diversity, and joining recombination. Our data indicate that RAG activity during T-cell receptor maturation induces a site-specific integration of AAV genomes and opens new therapeutic avenues for achieving long-term AAV-mediated gene transfer in dividing cells.
Introduction
Adeno-associated virus (AAV) vectors can efficiently transduce therapeutic transgenes into millions of somatic cells directly within patients’ organs via a single administration.1 The transgene is expressed from the AAV genome, which mainly persists in an episomal state in the nucleus of transduced cells. For this reason, AAV-based gene therapy (GT) applications have been successfully exploited in clinical trials to treat coagulation disorders, inherited blindness, and neurodegenerative diseases.1,2 Although AAV has been considered an episomal vector, we and others have previously reported that integration of fragmented or full-length AAV DNA occurs within the genome of host cells, such as hepatocytes, fibroblasts, muscle cells, and neuron cells, in both human and animal models (integration frequency: 0.1%-3%).3-10 Hotspots of AAV integrations are genomic sites with palindromic sequences, CpG islands, transcriptional start sites (TSSs), and active genes.3-13 Vector integrations are often characterized by inverted terminal repeat (ITR) deletions and other forms of vector rearrangements, potentially producing chromosomal rearrangements and deletions at the site of insertion.4,5,10,14 As AAV vectors do not encode for proteins required for integration, the entire mechanism is dependent on host factors. It has been shown that double-strand breaks (DSBs) or nicks in the host cell genome favor AAV integration via homologous and nonhomologous mechanisms of insertions.14 The availability of free chromosomal ends that can be joined with the AAV genome suggests that nuclear proteins involved in DNA damage response have a key role in regulating this process.14 Although preclinical and clinical studies aimed at addressing the safety of AAV integrations have shown a good safety profile without any adverse event linked to AAV integration reported in clinical trials,15-18 the finding that AAV is able to integrate in the host cell genome poses safety concerns for GT application. A single IV AAV administration may easily result in the transduction of hundreds of millions of cells, thus leading to a relevant number of AAV insertions. These insertions may end up being genotoxic in the long term, as shown in several preclinical safety studies, in which animals treated with high AAV doses developed hepatocellular carcinoma or clonal hepatocellular expansions after insertional activation of oncogenes, suggesting that insertional mutagenesis could be a safety concern for AAV GT applications.2,7,19-26
From a positive perspective, vector integrations guarantee that transgene expression will not decline because of tissue proliferation and that the correction can be transmitted to the cell progeny. The ability of AAV genomes to integrate at DSBs that are artificially induced via gene editing procedures has been successfully exploited to achieve stable genetic modification in GT applications for a wide variety of disorders, both in vivo and ex vivo.27-29 Targeting transgene insertions toward specific genomic sites can alleviate the risk of insertional mutagenesis, thereby increasing the safety of the GT protocol. Furthermore, in many AAV-based gene editing applications, the therapeutic benefit was conveyed by a selective advantage acquired by the transduced cells, as demonstrated in the preclinical model of liver-directed GT for hereditary tyrosinemia type1,5,30 alpha1 antitrypsin deficiency,31 Crigler-Najjar syndrome,32,33 and methylmalonic acidemia.34 We recently showed that AAV integration can provide therapeutic benefit even in a nonediting context of GT for T-cell immunodeficiency.35 In this work, AAV8 particles expressing Zeta-chain–associated protein kinase 70 (ZAP70) transgene were intrathymically injected into Zap70-deficient infant mice, promoting the development of functional T cells that persist for more than 40 weeks after treatment, associated with the rapid reconstitution of the thymic medulla.35 The long-term transgene expression and the maintenance of vector copy number observed in vivo in peripheral T lymphocytes and ex vivo after T-cell receptor (TCR) stimulation strongly suggested that T cells harbored integrated vector copy. Here, we characterized the AAV integration profile in mature lymph nodes (LNs), spleens (SPLs), and livers (LIVs) of ZAP70-treated mice as well as in an MeCP2 deficiency mouse model. In lymphoid tissues of intrathymically injected mice, clusters of AAV integrations were specifically detected in TCR genes at DNA breaks produced by the recombination activating genes (RAGs) enzyme during the variable (V), diversity (D), and joining (J) [V(D)J] recombination. These integrations resulted in the development of functional T lymphocytes and are likely responsible for their persistence for months after treatment, suggesting the important interest of this new targeting approach for GT applications.
Material and methods
Evaluation of vector genome quantity per diploid genome via qPCR
Vector genome copy number per diploid genome was assessed as previously described.35,36 Briefly, total DNA was extracted, and vector genome DNAs and diploid genomes were measured through the quantification of bovine growth hormone (BGH)-polyadenylation (pA) and endogenous albumin, respectively, via quantitative polymerase chain reaction (qPCR) using the Premix Ex Taq kit (Takara). For each qPCR, cycle threshold values were compared with those obtained using dilutions of plasmids harboring either the BGH-pA or albumin genes; results are expressed as vector genome (vg) per diploid genome.
Retrieval of AAV integration sites: library preparation and bioinformatic analyses via RAAVioli pipeline
To assess AAV integration, we adopted a sonication-based linker-mediated PCR method (SLiM), as previously described. Briefly, genomic DNA was sheared using a Covaris E220 Ultrasonicator (Covaris Inc),37,38 generating fragments with a target size of 1000 bp. The fragmented DNA was subjected to end repair, 3′ adenylation, and ligation (NEBNext Ultra DNA Library Prep Kit for Illumina, New England Biolabs) to custom linker cassettes (LCs; Integrated DNA Technologies). LC sequences contained an 8-nucleotide barcode for sample identification. Ligation products were subjected to 35 cycles of exponential PCR with primers (available upon request) complementary to different regions of the AAV genomes (supplemental Figure 2A; available on the Blood website) and to the LC. For each set of AAV-specific primers, the procedure was performed using ∼50 to 100 ng of sheared DNA. Then, 10 additional PCR cycles were run to include the sequences required for sequencing and a second 8-nucleotide DNA barcode. PCR products were quantified via qPCR using the Kapa Biosystems Library Quantification Kit for Illumina, following the manufacturer’s instructions. qPCR was performed in triplicates on each PCR product diluted 10:3, and the concentrations were calculated by plotting the average cycle threshold values against the provided standard curve. Finally, the amplification products were sequenced using Illumina Next/Novaseq platforms (Illumina).
After sequencing, a dedicated bioinformatics pipeline, recombinant adeno-associated vector integration analysis (RAAVioli), was developed to analyze the amplified sequences for integration site identification. Specific details of the pipelines will be reported in a follow-up methodological paper. A quality checks of input sequences was performed with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/, version v0.11.80), then adapter and PhiX reads were removed using Flexbar (version 3.5),39 whereas the first 12 random nucleotides were eliminated with Trimmomatic (version 0.39).40 The remaining portion of the sequences were aligned to the AAV genome using Burrows-Wheeler aligner–maximal exact match (with parameters -k 16 -r 1 -M -v 1 -T 15).41 Among the reads correctly aligned to the AAV sequences, those starting with the sequence of the last PCR primers used for amplification plus 8 to 10 vector-specific nucleotides were selected. Next, selected reads were aligned on a hybrid genome composed of both the AAV and mouse genome (release mm9 downloaded from University of California [Santa Cruz] Genome Browser website). A custom Python software was used to identify integration loci and vector rearrangements using concise idiosyncratic gapped alignment report string (supplemental Figure 3). Reads characterized by the same AAV/human genome breakpoint and the same number and type of AAV rearrangements were considered identical integration sites (ISs). Potential indels between the AAV junction and genomic locus were considered to distinguish 1 or 2 ISs: 2 reads from the same PCR reaction were assigned to the same IS if both alignments on human genome and the AAV junctions are aligned within a window of 20 bases. The abundance of each IS was measured by counting, for the same vector/cell genome junctions (IS), the number of different DNA fragments containing a genomic segment variable in size depending on the shear site position, and that will be unique for each different cell genome contained in the starting cell population. Therefore, the number of different shear sites assigned to an IS will be proportional to the initial number of contributing cells in the population studied, thus the clonal abundance of each IS in the starting sample does not consider the biases introduced with PCR amplification. Only integration events identified by at least 2 independent DNA fragments were considered, in the analyses, to remove potential PCR artifacts. The resulting table with all independent AAV IS reads is available in the SRA BioProject (PRJNA832955).
Identification of enriched motif
To find most enriched motifs in the sequences surrounding (± 50 nucleotides) the IS, we used multiple enriched motifs (EMs) for motif elicitation algorithm42 with the following options: motif site distribution: 0 or 1 site per sequence; maximum number of motifs: 1000; motif E-value threshold: 0.1; minimum motif width: 6; maximum motif width: 9; minimum sites per motif: 2; bias on number of sites: 0.8; sequence prior: simple Dirichlet; sequence prior strength: 0.01; EM starting point fuzz: 0.5; EM maximum iterations: 50; EM improvement threshold: 0.001; trim gap open cost: 11; trim gap extend cost: 1; and end gap treatment: same cost as other gaps.
Databases adopted for annotation of integration sites
Reference Sequences (RefSeq), CpG, TSS, RAG signal sequence (RSS), TCR, and END-seq analysis were performed using bedtools closest43 using the databases obtained as described below. RefSeq, TSS, and CpG databases were downloaded from Table Browser using the track RefSeq Genes. RSS database was obtained from https://www.itb.cnr.it/rss/index.html. TCR databases were obtained from the international ImMunoGeneTics information system for immunoglobulins or antibodies (IMGT) website:
TRA locus (Mus musculus|C57BL/6J)
TRB locus (Mus musculus|C57BL/6J)
TRD locus (Mus musculus|C57BL/6J)
TRG locus (Mus musculus|C57BL/6J)
DSB END-seq database was obtained from Canela et al,51 and are reported in supplemental Table 7. TCRs and END-seq databases were converted from mm10 coordinates to mm9 coordinates using genome liftover (https://genome.ucsc.edu/cgi-bin/hgLiftOver).
Statistical analysis
Statistical analyses were performed using GraphPad Prism 8.0 and R software.
Results
Identification and characterization of AAV integration sites
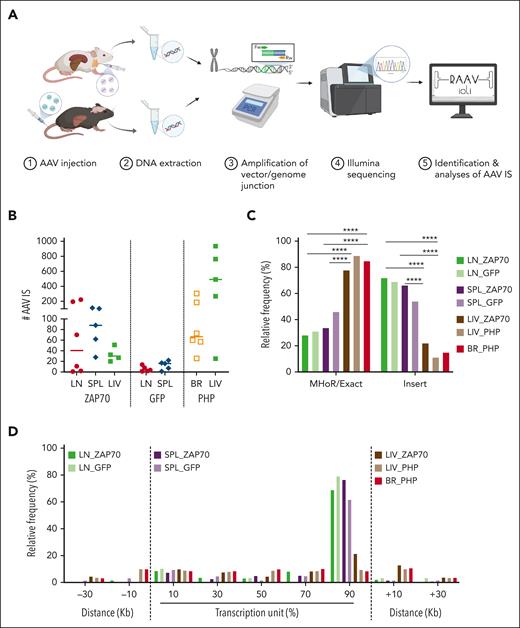
AAV ISs were retrieved from the genomic DNA collected from LNs (N = 7) and SPL tissues (N = 5) of Zap70-deficient mice intrathymically injected with AAV8 expressing the therapeutic transgene. Furthermore, because AAV particles can spread outside the thymus, leading to the transduction of other tissues, we included IS analyses of genomic DNA purified from LIV tissues (N = 3) that harbored AAV copies per diploid genome ranging from 0.001 to 1, between 3 and 43 weeks after treatment (Figure 1A; supplemental Figure 1). AAV ISs were also identified in LN (N = 5) and SPL tissues (N = 5) of wild-type (WT) mice intrathymically injected with a green fluorescent protein (GFP)-expressing AAV8 to overcome any selection bias conferred by the expression of the ectopic ZAP70 gene expression in T lymphocytes35 (supplemental Table 1). For the independent control, AAV ISs were retrieved from a preclinical model of GT for Rett syndrome, a severe neurological disorder caused by loss-of-function mutations in MeCP2, which encodes for a global chromatin regulator highly expressed in neurons. In this context, Mecp2-deficient mice were systemically administered with Mecp2-expressing AAV engineered with the PHP.eB capsid, which enables viral particles to permeate the blood-brain barrier, allowing the transduction of neurons and glia cells with high efficiency.44,45 Brain (BR) tissues were collected from Mecp2-deficient mice after treatment with 10-fold increasing doses of AAV-PHP.eB-Mecp2 (1 ×1010 vg, 1 × 1011 vg, and 1 × 1012 vg per mouse, N = 6 or 2 mice per dose; Figure 1A).45 Because PHP.eB viral particles can also transduce peripheral organs, AAV ISs were also retrieved from the LIV of Mecp2-deficient mice treated with PHP-AAV expressing GFP or the therapeutic transgene (N = 5; supplemental Table 1). Because Mecp2 expression did not alter or favor the selection of transduced hepatocytes, ISs in these groups of mice were merged (refer to supplemental Table 1 for information regarding the vector dose, vector copy number, and time points of tissue collection).
Retrieval of AAV ISs. (A) Schema of the experimental strategy. LN cells, SPL, and LIV tissues were collected from Zap70-deficient mice treated with AAV8. BR and LIV tissues were collected from Mecp2-deficient mice systemically injected with AAV-PHP. DNA was purified from the collected tissues and SLiM-PCR was performed to amplify vector/host-genome junction. PCR amplicons were assembled into libraries, sequenced, and reads were analyzed using RAAVIoli, a bioinformatics pipeline tailored to identify AAV ISs. (B) The numbers of AAV ISs retrieved from LN-derived mature T cells from LN, SPL, LIV, and BR tissues from the different groups of treated mice are presented with the median indicated as a colored line. (C) Relative frequency (indicated as percentage) of AAV ISs characterized by either precise homology breakpoints (Exact) and microhomology regions (MHoRs) between the vector and the host chromosomal sequences or random nucleotide insertions (insert). (D) Integration site distribution within gene bodies and their surrounding genomic regions. Each gene interval was quantified from the TSS up to the end of its coding region; this interval is considered as 100% and then normalized in bins of 20%. The surrounding genomic regions are divided in intervals of 20 kb. In panel C, statistical analyses were performed using a Fisher exact test ∗∗∗∗ P < .0001; see supplemental Table 2 for detailed statistical comparison.
Retrieval of AAV ISs. (A) Schema of the experimental strategy. LN cells, SPL, and LIV tissues were collected from Zap70-deficient mice treated with AAV8. BR and LIV tissues were collected from Mecp2-deficient mice systemically injected with AAV-PHP. DNA was purified from the collected tissues and SLiM-PCR was performed to amplify vector/host-genome junction. PCR amplicons were assembled into libraries, sequenced, and reads were analyzed using RAAVIoli, a bioinformatics pipeline tailored to identify AAV ISs. (B) The numbers of AAV ISs retrieved from LN-derived mature T cells from LN, SPL, LIV, and BR tissues from the different groups of treated mice are presented with the median indicated as a colored line. (C) Relative frequency (indicated as percentage) of AAV ISs characterized by either precise homology breakpoints (Exact) and microhomology regions (MHoRs) between the vector and the host chromosomal sequences or random nucleotide insertions (insert). (D) Integration site distribution within gene bodies and their surrounding genomic regions. Each gene interval was quantified from the TSS up to the end of its coding region; this interval is considered as 100% and then normalized in bins of 20%. The surrounding genomic regions are divided in intervals of 20 kb. In panel C, statistical analyses were performed using a Fisher exact test ∗∗∗∗ P < .0001; see supplemental Table 2 for detailed statistical comparison.
The retrieval of AAV IS was performed via SLiM-PCR, a technique used for clonal tracking and safety studies in hematopoietic stem cell GT clinical trials for rare diseases and cancer immunotherapy, based on lentivirus vectors, gamma retrovirus vectors, and transposons.37,38,46,47 Briefly, sonicated DNA was ligated to a barcoded LC and subjected to PCR amplification using primer sets specifically designed to cover different portions of the AAV genome (ie, ITRs, internal promoter, and the poly-A sequence), maximizing the possibility of amplifying vector/host-genome junctions that may originate from different fragmented forms of the integrated vector (supplemental Figure 2A). The resulting PCR products were sequenced and analyzed by an ad hoc bioinformatics pipeline tailored to identify AAV IS and are referred to as RAAVioli. RAAVioli filtered sequencing reads for quality, removed adapters, barcode sequences, and linker sequences and aligned the remaining portion to the AAV and mouse genome (Figure 1A; supplemental Figure 2B). Only sequencing reads containing AAV sequences downstream to the previous oligonucleotide used for the amplification (minimum length of 10 nucleotides) were selected for further analyses. Although most of the sequencing reads aligned on the AAV genome alone, 12% of the reads aligned on the mouse genome, revealing that they potentially contained a vector/host-genome junction (supplemental Table 1). The precise position of the IS was defined at the homology breakpoint between AAV and mouse genomic sequences (see “Material and Methods”). After collapsing chimeric reads with the same IS and homology structure, we identified 972, 394, and 131 nonredundant IS from the LN, SPL, and LIV of Zap70-deficient mice. Here, these IS data sets will be referred to as LN_ZAP70, SPL_ZAP70, and LIV_ZAP70, respectively. Fewer AAV ISs were obtained in WT mice treated with the GFP-expressing AAV8 than in Zap70-deficient mice: 29 and 63 IS were identified in the LN and SPL, respectively (hereinafter referred to as LN_GFP, SPL_GFP data sets, or GFP-treated mice). From Mecp2-deficient injected mice, 705 and 2488 nonredundant IS were retrieved from the brain and liver (BR_PHP and LIV-PHP data sets, respectively; Figure 1B; supplemental Table 1; supplemental Figure 2C-D). A significant and positive correlation was observed between the number of ISs and the doses of AAV vector injected in Mecp2-deficient mice (supplemental Figure 2E; P = .011 using Pearson correlation test).
By aligning the flanking sequences of the AAV vector-host-genome junctions, we observed that 36% ± 5.4% of the ISs from the different data sets presented a precise homology breakpoint (±1 nucleotides), which distinguished the nucleotide portion aligning to the AAV genome from that aligning on the mouse genome (Figure 1C; supplemental Figure 3A-B). In contrast, 64% ± 5.4% of the ISs from the different data sets were characterized by the presence of short stretches of nucleotides (up to 30 nucleotides) that could be equally mapped to the provirus and the mouse genome or had a random origin (supplemental Figure 3B). These data are in agreement with previous research demonstrating the presence of microhomology areas between the vector and chromosomal sequences (referred to as MHoR) as well as random nucleotide insertions (referred to as insertion) at AAV vector/host-genome junctions.5,6,12,13,26 Notably though, a comparison of the frequency of these events among the different IS data sets revealed that, independently from the Zap70 or WT genetic background and vector type, the LN- and SPL-derived data sets presented a significantly higher frequency of IS with random nucleotide insertions at the AAV vector/host-genome junctions compared with the LIV- and BR-derived IS data sets obtained from Zap70- and Mecp2-deficient mice (Figure 1C; supplemental Table 2; P < .0001 usingFisher exact test; see supplemental Table 2 for detailed statistical comparison). The increased frequency of short stretches of random nucleotides in the LN- and SPL-derived IS data sets compared with the others suggested the presence of a differentially regulated biological mechanism controlling the AAV integration processing in those tissues.
TCR genes are the most frequently targeted genes in LN-derived mature T cells in Zap70-deficient mice
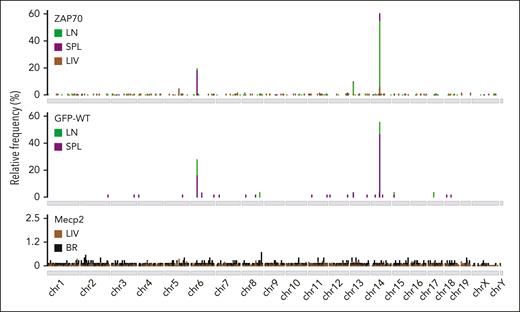
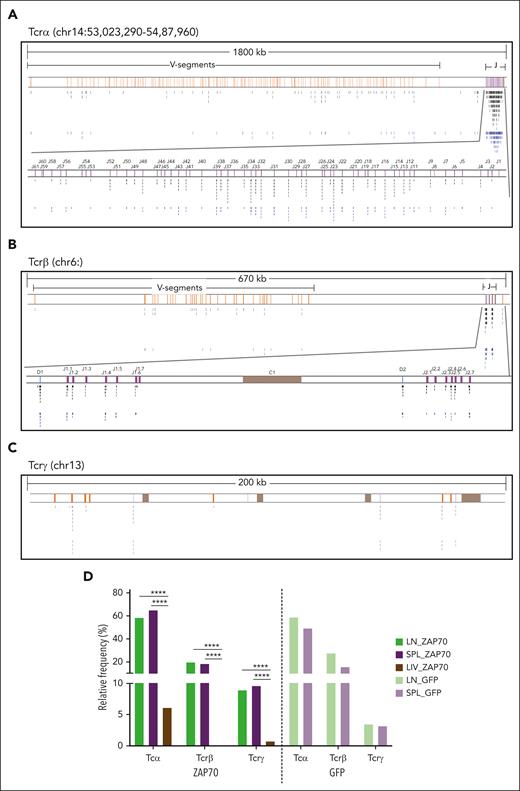
Analyzing the distribution of AAV ISs within the mouse genome, we observed that, independently of the mouse background or the AAV type used, the LIV and BR data sets displayed the typical preference of AAV IS to integrate close to CpG islands and TSSs (supplemental Figure 4A-B). In marked contrast, the LN and SPL data sets showed a peculiar distribution in CpG and TSSs, with a significantly higher integration frequency within gene bodies (91.4% ± 3% on average), compared with those observed in the LIV and BR IS data sets (47.3% ± 2% on average; see supplemental Table 3 for detailed statistical comparison; Figure 1D). Although ISs were sparse within the mouse genome in the LIV and BR data sets, 3 clusters of AAV IS were identified at chromosomes 6, 13, and 14 in the LN and SPL data sets of intrathymically injected mice (Figure 2A), reaching more than 84% of total integration events. Indeed, 567 (58.3%) and 257 (65.2%) different IS in the LN and SPL data sets, respectively, from the Zap70-treated mice, clustered in a genomic region of ∼1800 kb at chromosome 14 where the Tcrα/Tcrδ locus is annotated (Figure 3A; supplemental Table 4). Similarly, 17 (58.6%) and 31 (49.2%) IS from the LN and SPL data sets, respectively, from the GFP-treated mice targeted the Tcrα/Tcrδ locus (supplemental Figure 5A). A second and third cluster of AAV ISs spanned over 2 genomic regions of ∼670 and ∼200 kb at chromosomes 6 and 13 where Tcrβ and Tcrγ are located, respectively (Figure 3B-C; supplemental Figure 5B-C; supplemental Table 4). The Tcrβ locus was targeted by 192 (19.8%) and 73 (18.5%) different ISs in the LN and SPL data sets from the ZAP70-treated mice, respectively, and by 8 (27.6%) and 10 (15.9%) ISs in the LN and SPL data sets of the GFP-treated mice, respectively. Finally, the Tcrγ locus was targeted by 87 (9.0%) and 38 (9.6%) IS of the LN and SPL data sets, respectively, from the ZAP70-treated mice, and by 1 and 2 ISs of the LN and SPL data sets, respectively, from the GFP-treated mice (supplemental Table 4). Among the other targeted genes in the ZAP70 data set, we found 20 AAV IS targeting the IgH locus (chr12, 15 in LN, 5 in SPL) (Figure 3A; supplemental Figure 5D). A single AAV IS in IgH and 2 IS in IgK (chr6) were identified in the SPL-derived data set of GFP-treated mice. In the LIV data set of ZAP70-treated mice, TCRα was the most targeted gene (N = 8, 6.2%), followed by albumin gene (Alb, N = 4, 3.1%) (supplemental Table 4). Although TCRα was targeted genes by different IS, the targeting frequency of the TCR loci was significantly reduced compared with that identified in the LN and SPL data sets of ZAP70-treated mice (Figure 3E; P < .0001 using Fisher exact test). Of note, 47 ISs in the ZAP70 data set were retrieved in different tissues from the same mouse: 9 ISs from mouse 101 and 37 ISs from mouse 111 were found in LN and SPL, whereas 1 IS from mouse 103 was retrieved in the LIV and SPL, suggesting the trafficking of transduced T cells to different organs. Most of these shared IS-targeted TCR loci (38 Tcrα, 6 Tcrβ and 1 Tcrγ, N = 45, 96% frequency). In contrast, no IS in TCR loci were identified in the LIV and BR IS data sets of Mecp2-deficient mice. Neuron- and LIV-specific expressed genes, such as Ncam2, Prcc2b for the BR data set (N = 3) and Alb for the LIV (N = 9), were the most targeted (supplemental Table 4). Furthermore, 3 ISs in immunoglobulin H (IgH), 1 IS in IgK, and 1 IS in IgL (chr16:19061845-19260937) were found in the LIV and BR data sets of PHP-treated Mecp2-deficient mice.
Cluster of AAV ISs within TCR genes. Genome-wide distribution of AAV ISs in the different tissues of ZAP70-deficient (ZAP70), WT mice, and Mecp2-deficient mice (Mecp2) intrathymically injected with AAV: LN, SPL, LIV, and BR are shown as indicated. Most of AAV integrations in the LN and SPL data sets from intrathymically injected mice were detected at chr6, 13, and 14.
Cluster of AAV ISs within TCR genes. Genome-wide distribution of AAV ISs in the different tissues of ZAP70-deficient (ZAP70), WT mice, and Mecp2-deficient mice (Mecp2) intrathymically injected with AAV: LN, SPL, LIV, and BR are shown as indicated. Most of AAV integrations in the LN and SPL data sets from intrathymically injected mice were detected at chr6, 13, and 14.
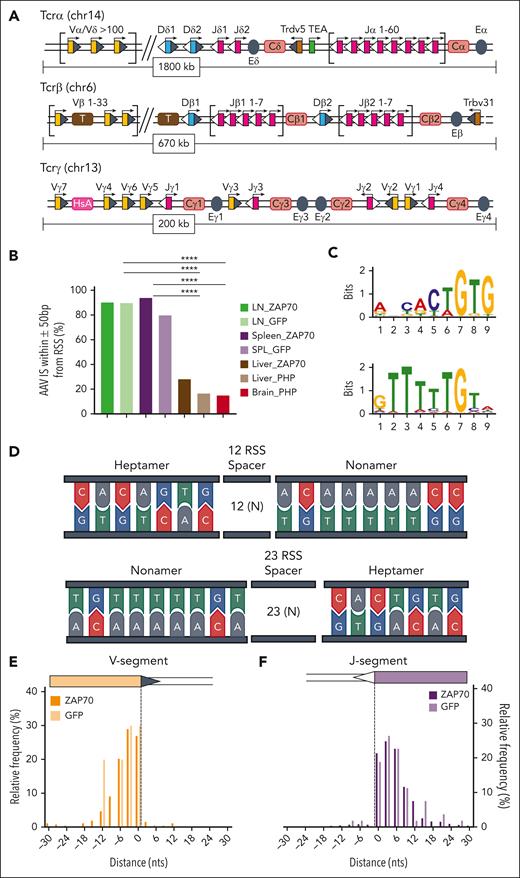
Distribution of AAV ISs within Tcr loci. (A-C) Genomic distribution of AAV IS in the LN and SPL data sets of ZAP70-treated mice and targeting TCR genes: TCRα (A), TCRβ (B), and TCRγ (C), as indicated. Genomic coordinates and scale are indicated in each panel. Black and blue lines indicate the position of the AAV IS from the LN (black) and SPL (blue) data sets. V, D, and J segments are indicated by yellow, blue, and purple rectangles, respectively. (C) Constant regions are represented as brown rectangles. Clusters of AAV ISs are identified in TCR genes, especially in the J-segment region, whose genomic area is enlarged below the Tcrα and Tcrβ loci; gene segment genomic coordinates were retrieved from IMGT, the international ImMunoGeneTics information system. (D) Gene targeting frequency (indicated as percentage) of Tcrα, Tcrβ, and Tcrγ in the different AAV ISs data sets as indicated.
Distribution of AAV ISs within Tcr loci. (A-C) Genomic distribution of AAV IS in the LN and SPL data sets of ZAP70-treated mice and targeting TCR genes: TCRα (A), TCRβ (B), and TCRγ (C), as indicated. Genomic coordinates and scale are indicated in each panel. Black and blue lines indicate the position of the AAV IS from the LN (black) and SPL (blue) data sets. V, D, and J segments are indicated by yellow, blue, and purple rectangles, respectively. (C) Constant regions are represented as brown rectangles. Clusters of AAV ISs are identified in TCR genes, especially in the J-segment region, whose genomic area is enlarged below the Tcrα and Tcrβ loci; gene segment genomic coordinates were retrieved from IMGT, the international ImMunoGeneTics information system. (D) Gene targeting frequency (indicated as percentage) of Tcrα, Tcrβ, and Tcrγ in the different AAV ISs data sets as indicated.
Overall, these data revealed a massive enrichment of AAV IS in TCR loci (α, β, and γ) in the LN and SPL of intrathymically transduced Zap70-deficient and WT mice.
AAV integrations occurred in genomic regions subjected to recombinase-mediated rearrangements
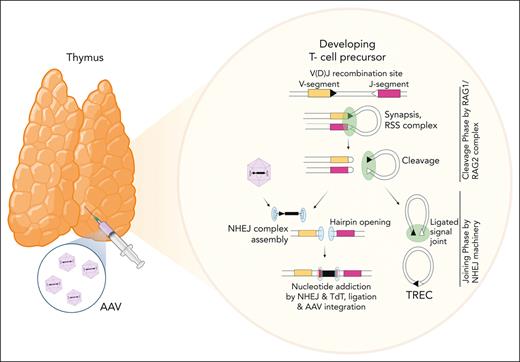
TCR genes are composed of multiple dispersed gene segments, including the V(D)J segments that recombine during thymocyte development through a process known as V(D)J recombination. Recombination is directed by the lymphoid-specific recombinase enzyme (RAG, composed of RAG1 and RAG2) and ubiquitously expressed DNA repair proteins48,49 (Figure 4A; supplemental Figure 6). RAG enzymes create DSBs at recombination signal sequences (RSSs) that flank TCR V, D, and J gene segments, and these breaks are subsequently resolved by nonhomologous end joining (NHEJ) mechanisms. At the end of the process, a new rearranged gene configuration is produced, leading to the expression of a functional TCR.
AAV integrations occurred at RAG-induced DSBs. (A) Schematic representation of the organization of the murine Tcrα/Tcrδ, Tcrγ, and Tcrβ loci. Genomic proportions are not preserved. V, D, and J segments are indicated by yellow, blue, and purple rectangles, respectively. RSS are represented as triangles close to the V, D, and J gene segments: RSS with a 12-bp spacer are indicated as grey triangles, RSS with a 23-bp spacer are indicated as white triangles. TCR constant regions are indicated as Ca, Cb, Cg and Cd and represented as pink rectangles. The T early exon (TEA) in the TCRα locus is represented by a green rectangle. Trypsinogen genes in the TCRβ locus (T) are represented by brown rectangles. Enhancers are represented by grey circles and indicated as E. Germ line transcription is indicated by black arrows above each segment. (B) Frequency (indicated as percentage) of AAV ISs surrounded by candidate RSSs in a genomic window of 100 bp (± 50 bp). (C) Significant motifs identified by MEME algorithm within the LN and SPL data set of ZAP70-treated mice. (D) Schema of the conserved heptamer and nonamer sequence of RSS. (E, F) Distribution of AAV ISs at the 3′ region of V segments (E) and at the 5′ region of J segments (F). Statistical analyses in panel B were performed using the Fisher exact test. ∗∗∗∗P < .0001. chr, chromosome; MEME, multiple Em for motif elicitation.
AAV integrations occurred at RAG-induced DSBs. (A) Schematic representation of the organization of the murine Tcrα/Tcrδ, Tcrγ, and Tcrβ loci. Genomic proportions are not preserved. V, D, and J segments are indicated by yellow, blue, and purple rectangles, respectively. RSS are represented as triangles close to the V, D, and J gene segments: RSS with a 12-bp spacer are indicated as grey triangles, RSS with a 23-bp spacer are indicated as white triangles. TCR constant regions are indicated as Ca, Cb, Cg and Cd and represented as pink rectangles. The T early exon (TEA) in the TCRα locus is represented by a green rectangle. Trypsinogen genes in the TCRβ locus (T) are represented by brown rectangles. Enhancers are represented by grey circles and indicated as E. Germ line transcription is indicated by black arrows above each segment. (B) Frequency (indicated as percentage) of AAV ISs surrounded by candidate RSSs in a genomic window of 100 bp (± 50 bp). (C) Significant motifs identified by MEME algorithm within the LN and SPL data set of ZAP70-treated mice. (D) Schema of the conserved heptamer and nonamer sequence of RSS. (E, F) Distribution of AAV ISs at the 3′ region of V segments (E) and at the 5′ region of J segments (F). Statistical analyses in panel B were performed using the Fisher exact test. ∗∗∗∗P < .0001. chr, chromosome; MEME, multiple Em for motif elicitation.
The high level of clustering of AAV ISs in TCRs appeared to be the result of the trapping of the AAV genome at the DSBs produced by RAG enzymes during the T-cell maturation process. To confirm this hypothesis, we evaluated the presence of candidate RSS in the surrounding genomic region (± 50 nucleotides) of each AAV IS in the different data sets.50 These analyses revealed that a candidate RSS lies adjacent to AAV IS in more than 88% ± 3% of the AAV ISs derived from the LN and SPL data sets of intrathymically injected mice, whereas a significantly lower frequency (20.1% ± 4%) was observed in the LIV and BR data sets obtained from Zap70- and Mecp2-deficient mice (Figure 4B; P < .0001 using Fisher exact test; supplemental Figure 6; supplemental Table 5 for detailed statistical comparison).
Using the multiple Em for motif elicitation algorithm,42 we analyzed the mouse genomic sequences encompassing the vector integration sites (±50 bp) in the LN and SPL data sets and found that the 2 most significantly enriched motifs (maximum E-value <0.01) closely resembled the conserved 7 (heptamer) and 9 (nonamer) elements of RSS signals (Figure 4C-D). These RSS-like motifs were not found in the other IS data sets. Rather, this analysis revealed a significant enrichment for GC-rich and degenerated palindromic motifs (supplemental Figure 7). RSSs can be distinguished based on the 12- or 23-bp spacer lengths separating conserved heptamer and nonamer elements. The V(D)J recombination process is restricted to gene segments flanked by dissimilar RSSs following the 12 ÷ 23 rule, so that a V-to-D recombination occurs only when a 23-RSS that is downstream of a V segment is joined with a 12-RSS upstream of a D segment.48,49 A closer inspection of the distribution of the AAV IS within TCRs revealed that 94% and 92% of integration events of Zap70 and WT intrathymically injected mice clustered within the 3′ region of V segments and the 5′ region of J segments, hence in proximity of DSBs created by RAG enzymes recruitment to 12-RSS and 23-RSS substrates (Figure 4E-F; supplemental Figure 6; supplemental Table 5-6). Furthermore, 95.2% and 100% of the AAV ISs in the TCRα locus of Zap70-deficient and WT-injected mice, respectively, occurred in regions that were identified in mouse thymocyte via END-seq, a technology platform that allows the genome-wide identification of DSBs51 (supplemental Figure 6; supplemental Table 7). These data, therefore, confirm our hypothesis that AAV IS occurs in genomic regions that are subjected to RAG recombinase-mediated rearrangements.
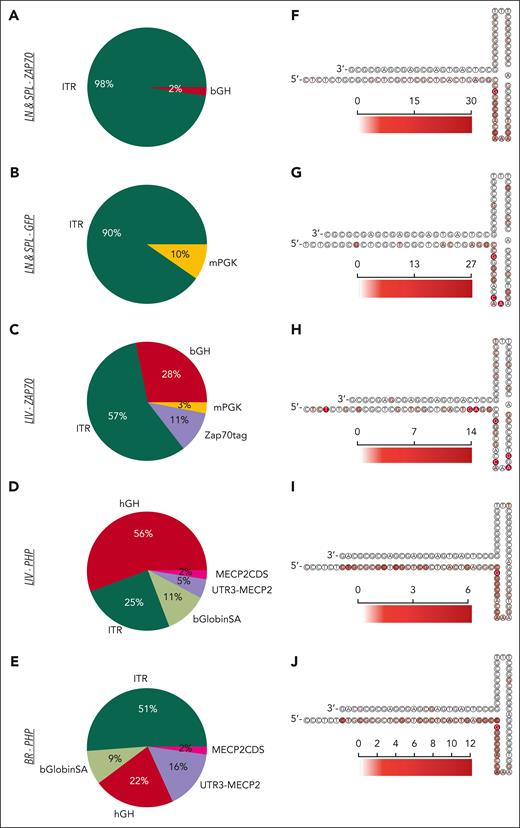
We then investigated the regions of the AAV genome that were involved in the integration events. In the LN and SPL data sets of intrathymically injected mice, more than 90% of integrations involved the ITRs (Figure 5A-B), whereas in the LIV and BR data sets of Zap70- and Mecp2-deficient mice, the poly-A, the transgene coding sequence, and the promoter were also identified at the junction breakpoints (Figure 5C-E). Finally, a more granular inspection identified some preferred nucleotide breakpoints within loop regions A and C of ITRs (Figure 5F-L), as previously reported.13,26,27
AAV genomic features at the vector/host-genome junction. (A-E) Pie charts indicating the frequency of AAV features found at the junction breakpoint between the integrated vector and the host genomic region for the different IS data sets as indicated. (F-J) Heatmap of the AAV 3′ITR secondary structure with the red scale indicating the frequency of AAV insertions occurring at the indicated nucleotide position for the different IS data sets. bGlobinSA, beta globin splice acceptor; CDS, coding sequence; hGH, human growth hormone; mPGK, murine phosphoglycerate kinase.
AAV genomic features at the vector/host-genome junction. (A-E) Pie charts indicating the frequency of AAV features found at the junction breakpoint between the integrated vector and the host genomic region for the different IS data sets as indicated. (F-J) Heatmap of the AAV 3′ITR secondary structure with the red scale indicating the frequency of AAV insertions occurring at the indicated nucleotide position for the different IS data sets. bGlobinSA, beta globin splice acceptor; CDS, coding sequence; hGH, human growth hormone; mPGK, murine phosphoglycerate kinase.
AAV integrations are stably integrated in the genome of proliferating T cells
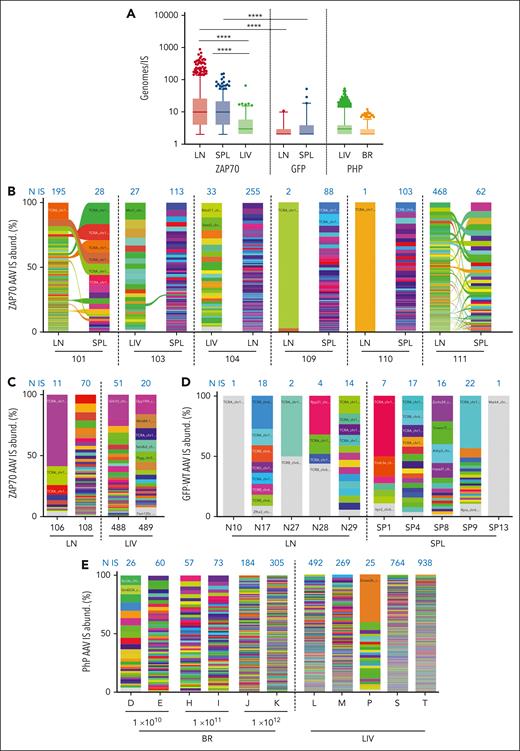
Our SLiM-PCR protocol was performed using sonicated genomic DNA material, which allowed for the abundance of each AAV IS to be estimated by counting the numbers of different genomic shear sites. Multiple cellular genomes in the LN and SPL data sets were associated with the same AAV IS, highlighting the stable inheritance of AAV IS by cell progeny after T-cell proliferation. These results confirm the maintenance of integrated AAV copies, as previously reported,35 and exclude the possibility that AAV IS occurred in T-cell receptor excision circles, episomal byproducts that originate from the excision of the delta-coding sequences during TCR recombination. Moreover, a significantly higher number of genomes per IS was observed in the LN and SPL data sets (average 31.8 ± 2.3 and 16.7 ± 1.0 genomes per IS in LNs and SPLs, respectively) than in the LIV data sets of ZAP70-treated mice (average 5 ± 0.6; P < .0001 using the two-tailed Mann-Whitney test; Figure 6A), likely reflecting the higher proliferative capacity of T cells as compared with LIV cells under physiological conditions. A significantly lower number of genomes was observed in the LN and SPL data sets of GFP-treated WT mice compared to ZAP70-treated mice, and this was probably because of the selective advantage conferred by the expression of the integrated therapeutic transgene in transduced T cells in the latter mice.
Absolute and relative abundance of AAV integration sites. (A) Cellular genomes observed for each IS (Genomes/IS) in the different IS data sets are presented as indicated. (B-E) Stacked bar plots showing the relative abundance of AAV IS retrieved in ZAP70-treated mice (B,C), in WT mice intrathymically injected with a GFP-expressing AAV (D), and in Mecp2-deficient mice (E). In each column, every integration is represented by a different color whose height is proportional to the number of genomes retrieved for that specific IS (percentage of IS abundance, y-axis). The number of unique IS identified in each mouse is indicated in blue above each column. Ribbons connect AAV ISs tracked between different tissues of the same mouse. In A and B, statistical analyses were performed by a two-tailed Mann-Whitney test. ∗∗∗∗P < .0001.
Absolute and relative abundance of AAV integration sites. (A) Cellular genomes observed for each IS (Genomes/IS) in the different IS data sets are presented as indicated. (B-E) Stacked bar plots showing the relative abundance of AAV IS retrieved in ZAP70-treated mice (B,C), in WT mice intrathymically injected with a GFP-expressing AAV (D), and in Mecp2-deficient mice (E). In each column, every integration is represented by a different color whose height is proportional to the number of genomes retrieved for that specific IS (percentage of IS abundance, y-axis). The number of unique IS identified in each mouse is indicated in blue above each column. Ribbons connect AAV ISs tracked between different tissues of the same mouse. In A and B, statistical analyses were performed by a two-tailed Mann-Whitney test. ∗∗∗∗P < .0001.
Finally, having estimated the number of genomes associated with each AAV IS, we also calculated the relative level of abundance for each IS as a percent of the total IS identified in each sample (Figure 6B-E). An overall polyclonal pattern was observed in the LIVs and BRs of Mecp2-deficient mice and in the LNs and SPLs from ZAP70-treated mice. However, in some cases, clones with a relatively high level of abundance can be identified because of the small number of IS recovered in that sample, such as in the LNs and SPLs of WT mice.
Discussion
Here, we characterize hundreds of AAV ISs from lymphocyte-derived tissues and compared their integration profile with those observed in LIV and BR tissues from 2 different preclinical models of GT. Notably, we found that >90% of the integrations in LNs and SPLs from intrathymically injected mice clustered within TCR genes (α, β, and γ) close to DNA breaks created by the enzymatic activity of RAGs during V(D)J recombination. Hence, to our knowledge, our work is the first to demonstrate that the intrathymic delivery of AAV leads to site-specific integration within TCR genes.
Most integration events occurred in Tcrα and β genes that are assembled together to form the TCRαβ heterodimer complex, presented at the surface of T cells late in the thymic developmental program.52 Recombination of Tcrδ and Tcrγ occurs during the double-negative stage of thymocyte development, and successful recombination of Tcrδ and Tcrγ promotes assembly of a γδ TCR. In contrast, successful recombination of TCRβ promotes its assembly with a pre-Tcrα, allowing the transition toward the double-positive stage, in which TCRα gene rearrangement occurs. Only the successful rearrangement of the TCRα chain results in the formation of a thymocyte with a mature TCRαβ heterodimer, and this cell can then differentiate into a single-positive thymocyte, the precursor of a mature peripheral T cell. In the treated ZAP70-deficient mice, the ectopic expression of ZAP70 from the AAV vector is required for effective TCRαβ heterodimer, signaling at the double-positive stage of thymocyte differentiation and subsequent T-cell differentiation and function.35,53 Hence, in this context, AAV integration into TCR genes affected neither the development nor the function of the transduced T cells. Peripheral gene-corrected T cells appeared as early as 3 weeks after vector administration, persisted for more than 10 months, and proliferated robustly after TCR stimulation, thus demonstrating a fully reconstituted TCR signaling cascade.35 Currently, it is not possible to determine whether vector integration occurred within the genomic allele encoding for the functional TCR gene. Nevertheless, because vector integration often occurrs within the first/last nucleotides of TCR gene segments, it is possible that its genomic presence may interfere with the successful rearrangement of the TCR chain, thus promoting the engagement of the sister allele. Irrespective, it is likely that the integration of the AAV-ZAP70 vector into the TCR loci was responsible for the long-term reconstitution of these animals.
The integration profiles obtained from the LN and SPL of WT mice injected with a GFP-expressing AAV are similar to those obtained in the LN and SPL of Zap70-deficient mice, in which AAV ISs mainly targeted TCR loci at RSS site of V(D)J gene segments (92% in ZAP70-treated mice and 85% in WT-treated mice). These data confirmed our finding that intrathymic injection favored the trapping of the AAV genome at RAG-induced DSBs and now show that this integration is independent of the role of the transgene in the T-cell differentiation process. The limited number of AAV ISs retrieved in WT mice compared with Zap70-deficient ones is due to the absence of a selection advantage conferred by the expression of the integrated transgene, which promotes the development and function of gene-corrected cells. In addition, in physiological conditions, nontransduced T cells continuously exit the thymus after differentiation.
Few AAV ISs targeting the genomic region proximal to V(D)J gene segments of IgH, IgK, and IgL genes (B-cell receptor loci) were identified in the different IS data sets. These integration events occurred exactly at the sites where the RAG recombinase complex mediates the DNA break for the rearrangement of Ig genes, suggesting that AAV can transduce B cells in the bone marrow during B-cell receptor recombination independently from the route of vector administration and the vector capsid, albeit at a very low frequency. The lower frequency of integration events in Ig loci compared with that in TCRs can be explained by the low frequency of B cells in the thymus and the lack of a selective advantage conferred by Zap70 expression in B cells. These findings are also consistent with our previously published data, in which low levels of B and myeloid cells expressing the therapeutic transgene were detected in AAV8–ZAP-70–treated mice during the 40-week follow-up period.35
In contrast to the LN- and SPL-derived data sets of intrathymically injected mice, AAV ISs in the LIV and BR data sets targeted highly expressed genes. Indeed, albumin was among the most targeted genes in the LIV data set of Zap70- and Mecp2-deficient mice, whereas Ncam2 and Pbbrc2 were the main targets in the BR of Mecp2-deficient mice. Preferential targeting of AAV ISs in genes with high expression levels can be explained by the higher DNA damage associated with high transcription levels, which may promote the occurrence of DNA breaks, thus favoring AAV integration.54,55 This mechanism was independent of the route of vector administration, the vector capsid, or the genetic background of the treated mice. In contrast, the presence of some IS targeting TCR loci in the LIV of ZAP70-treated mice is likely due to the trafficking of transduced T cells, as demonstrated by the detection of shared IS between different tissues of the same treated mouse.
The higher frequency of ITR sequences at the junction breakpoint in the LN and SPL data sets is of interest, considering the results obtained from the LIV IS data set of Zap70-deficient mice treated with the same vector whose genomic regions were interrogated using the same PCR systems. Although lymphocytes possess several DSB repair pathways, including the canonical NHEJ, homologous recombination, and additional microhomology–mediated pathways, RAG-initiated DSBs are almost exclusively fixed via NHEJ mechanisms that offer an optimal repair pathway allowing for TCR gene diversification.52,56 NHEJ is a rapid and efficient way of repairing DSBs because it involves the identification and ligation of the 2 DNA ends without searching for homology. It is usually restricted to the G1 phase of the cell cycle, which is when V(D)J recombination occurs. Furthermore, the peculiarity of DSB ends generated by the RAG nuclease, the protection of DNA ends against resection, and microhomology–directed repair are all factors that strongly promote repair via NHEJ rather than homologous recombination. It is likely that in this cell-specific setting, the binding of DNA damage response proteins, such as Ku70/80 heterodimer kinase (Ku), DNA-dependent protein kinase (DNA-PKs), ataxia-telangiectasia mutated kinase (ATM), and the Mre11-Rad5-Nbs1 (MRN) complex (MRE11, RAD50, and NBS1), on genomic DNA and on the free DNA ends of the vector ITRs may facilitate the joining and repair by NHEJ.52,56 The higher frequency of insertion of random nucleotides between the vector and chromosomal sequence in the amplified reads obtained from the LN and SPL data sets is likely due to the recruitment of the terminal deoxynucleotidyl transferase during V(D)J recombination, promoting template-independent and -dependent synthesis before ligation of the genomic and vector ITR ends. However, in the LIV and BR data sets, integration events can occur at DBSs and single-strand breaks, taking advantage of other DNA repair pathways that may involve other regions of the vector backbone. This is highlighted by the higher frequency of MHoR between the vector and chromosomal sequences in the ISs retrieved from the BR and LIV subsets as compared with the LN and SPL data sets.
As peripheral gene-corrected T cells can persist over a long period,35,36 this new targeting approach may open new therapeutic avenues, achieving long-term AAV-mediated gene transfer in dividing T cells. Hence, intrathymic AAV gene transfer might represent a novel approach for the treatment of primary immunodeficiencies, either alone for diseases that selectively affect T-cell development (such as CD3 or CD45 deficiencies) or in combination with hematopoeitic stem cell–based GT application for the treatment of other primary immunodeficiencies affecting multiple hematopoietic lineages (such as γc deficiency, JAK3 deficiency, etc). Intrathymic AAV-mediated gene transfer will promote a fast restoration of the thymic architecture and a rapid T-cell reconstitution, whereas the hematopoeitic stem cell correction will allow the restoration of all other hematopoietic lineages. In addition, the targeting of thymocytes with an AAV gene construct can be envisaged for immune-therapy applications redirecting T-cell activity against a specific antigen via the introduction of a new antitumor TCR or chimeric antigen receptor. From a safety standpoint, the genomic targeting of AAV vectors within TCR genes limits the risk of oncogenic transformation associated with the use of randomly integrated vectors. Furthermore, the selection processes occurring in the thymus during the T-cell maturation process may favor the deletion of transduced T-cell clones expressing dysfunctional/autoreactive TCRs, thus increasing the safety of the genetic modification and defining TCR genes as safe harbor loci for GT application. Finally, it would be interesting to assess the potential of intrathymic AAV-mediated gene transfer for the treatment of enzyme deficiencies, such as mucopolysaccharidosis type I, Hurler variant (MPSIH) or adenosine deaminase (ADA) deficiency, caused by a deficiency in the production of a secreted factor. In this condition, circulating transduced T cells could serve as a source of the factor of interest.
In conclusion, our results demonstrate that the intrathymic delivery of AAV is a conditioning-free approach of GT that induces a site-specific integration within DNA breaks created by the enzymatic activity of RAGs during V(D)J recombination. Because peripheral gene-corrected T cells can persist over a long period,35,36 this new targeting approach exploiting a naturally occurring biological process can open therapeutic avenues for achieving long-term AAV-mediated gene transfer in dividing T cells.
Acknowledgments
The authors thank all members of the Montini lab for their technical help and support.
This work was supported by Telethon Foundation TGT16B01 and TGT16B03 (E.M.); and Telethon #1350 (V.B.); AFM-Téléthon grants 22487 and 23743 (V.S.Z. and N.T.). M.P. was supported by AFM-Téléthon and the FRM.
Authorship
Contribution: A.C. and C.C. developed bioinformatics analyses, interpreted data, and wrote the manuscript; G.S., L.R., S. E., F.B., and A.A. provided great technical support; M.P. and M.G. performed the GT experiments on Zap70-deficient mice; M.L, S.G., and V.B. performed the GT experiments on Mecp2-deficient-mice; O.A., N.T., and V.S.Z. supervised gene therapy experiments on Zap70-deficient mice and N.T. and V.S.Z. contributed to writing of the manuscript; E.M. revised the work, interpreted data, and provided fruitful discussion; D.C. conceived and supervised the project, interpreted data, supervised research, wrote the manuscript, and coordinated the work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Daniela Cesana, San Raffaele Telethon Institute for Gene Therapy (Sr-Tiget), Via Olgettina 58, Milan 20131, Italy; e-mail: cesana.daniela@hsr.it.
References
Author notes
Sequencing data have been uploaded in NCBI Sequencing Read Archive BioProject (accession number PRJNA832955).
All original code has been deposited in GitHub (https://github.com/calabrialab/Code_AAV_IS_intrathymus).
Integration site matrixes for LN, LIV, and BR datasets are provided in supplemental Table 6; RSS, TCR, and END-seq annotations for each IS are available in supplemental Table 7.
All relevant data are included in the manuscript. Configuration settings to run RAAVioli are enclosed in this document.
Any additional data are available on request from the corresponding author, Daniela Cesana (cesana.daniela@hsr.it).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal