Key Points
CD53 is upregulated in HSCs in response to various inflammatory and proliferative stressors.
CD53 promotes HSC return to quiescence via promotion of the DREAM complex activity and downregulation of cycling-associated genes.
Abstract
The hematopoietic stem cell (HSC) cycle responds to inflammatory and other proliferative stressors; however, these cells must quickly return to quiescence to avoid exhaustion and maintain their functional integrity. The mechanisms that regulate this return to quiescence are not well understood. Here, we show that tetraspanin CD53 is markedly upregulated in HSCs in response to a variety of inflammatory and proliferative stimuli and that the loss of CD53 is associated with prolonged cycling and reduced HSC function in the context of inflammatory stress. Mechanistically, CD53 promotes the activity of the dimerization partner, RB-like, E2F, and multi-vulva class B (DREAM) transcriptional repressor complex, which downregulates genes associated with cycling and division. Proximity labeling and confocal fluorescence microscopy studies showed that CD53 interacts with DREAM-associated proteins, specifically promoting the interaction between Rbl2/p130 and its phosphatase protein phosphatase 2A (PP2A), effectively stabilizing p130 protein availability for DREAM binding. Together, these data identified a novel mechanism by which stressed HSCs resist cycling.
Introduction
Maintenance of quiescence is necessary for normal hematopoietic stem cell (HSC) function.1 Under homeostatic conditions, over 75% of HSCs reside in the G0 phase of the cell cycle.2,3 However, in response to stressful stimuli, HSCs are induced to proliferate and differentiate. Although this is important in the short term to supply effector cells necessary to resolve an acute insult, chronic exposure to inflammatory signals can lead to loss of sustained HSC function.4-6 Indeed, inflammatory inhibition of HSC function contributes to bone marrow (BM) suppression in patients with chronic infections, autoimmune disorders, and other BM failure syndromes.7
Multiple inflammatory stressors, including cytokines, such as granulocyte-colony stimulating factor (G-CSF), interferon 1 (IFN-1), and toll-like receptor (TLR) agonists, induce HSC proliferation.8-11 Notably, HSCs ultimately return to quiescence after exposure to these stressors, which is thought to help prevent HSC exhaustion during chronic inflammation. For example, exposure of HSCs to G-CSF or IFN-1 causes a transient burst of cycling over the initial days of treatment, with return to quiescence by day 3.8,10 Various surface receptors, cell-cell interactions, and cell-intrinsic signaling pathways have been shown to promote HSC cycling, but the mechanisms responsible for returning to quiescence are understudied.5,8,12,13
Herein, we found that CD53 expression was markedly upregulated in HSCs in response to various stressors, thereby promoting HSC quiescence and function. CD53 is a member of the tetraspanin family of transmembrane proteins that organize protein interaction networks on the cell membrane and regulate processes including cellular migration, adhesion, and signaling.14,15 We previously reported that CD53 regulates early B cell development via promotion of interleukin-7 receptor (IL-7R) signaling.16 CD53 has otherwise been implicated in the alteration of protein kinase C signaling, apoptosis, and proliferation in lymphocytes.17-19 Limited HSC data on CD53 have shown it to be asymmetrically segregated in HSCs, with CD53-enriched HSCs believed to be more stem-like20; however, there is yet to be a proposed mechanism to explain the association between CD53 and stem cell quality.
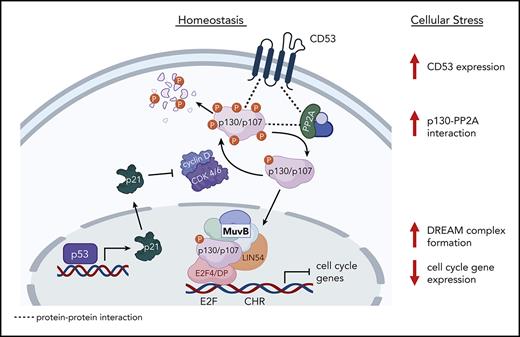
In this study, we showed that although Cd53−/− mice have largely normal HSCs under homeostatic conditions, the loss of CD53 is associated with a significant reduction in HSC function and prolonged cycling in response to stress. Consistent with this finding, transcriptomic sequencing revealed a significant upregulation of genes associated with cell cycling in G-CSF treated Cd53−/− HSCs compared with that in wild-type (WT) controls. Notably, most of these differentially expressed genes (DEGs) are targets of the dimerization partner, RB-like, E2F and multi-vulva class B (DREAM) complex, a transcriptional regulator that represses cell cycling in response to stress,21 suggesting that CD53 promotes DREAM-mediated quiescence in stressed HSCs. Indeed, promotion of DREAM binding using the CDK4/6 inhibitor palbociclib equalized the repopulating activity of G-CSF treated Cd53−/− and WT HSCs and restored nearly all DEGs. Biochemical analysis revealed that CD53 interacts with the DREAM complex subunit p130 (retinoblastoma-like 2; Rbl2), as well as the scaffold subunit, PP2A-Aα, of protein phosphatase 2A (PP2A), a phosphatase complex known to dephosphorylate p130, thus activating p130 and promoting cell quiescence.22 Furthermore, loss of CD53 results in a decrease in PP2A-Aα–p130 interactions in stressed HSCs, as well as a decrease in p130 protein levels. Together, these data support a novel mechanism whereby CD53 protects HSCs in the context of inflammatory stress by promoting DREAM complex activity and returning to quiescence.
Methods
Mice
Cd53−/− mice were generated on a C57BL/6J background as previously described16 and back-crossed at least twice (to C57BL/6J; Jackson Laboratory, Bar Harbor, ME) before experimental use. WT littermates were used as the controls. Sex- and age-matched mice were used for each experiment, in accordance with the guidelines of the Washington University Animal Studies Committee.
Flow cytometry
Peripheral blood was obtained via a cardiac puncture. BM hematopoietic cells were isolated by centrifugation of long bones at 6000 rpm for 3 minutes. Spleen cells were harvested by gentle crushing using a 100 μm strainer. The cells were processed for staining as previously described,23 and stained using the antibodies listed in supplemental Table 8 (available on the Blood website). Cell counts were determined using Hemavet HV950 (Drew Scientific of Erba Diagnostics, Miami Lake, FL) or Element HT5 (Heska Corporation, Loveland, CO). Stained cells were analyzed on a BD flow cytometer (BD Biosciences, San Jose, CA), Gallios flow cytometer (Beckman Coulter, Indianapolis, IN), Attune NxT flow cytometer (ThermoFisher, Santa Clara, CA), or NovoCyte flow cytometer (Agilent Technologies, San Diego, CA). The data were analyzed using FlowJo software (version 10.5.3; TreeStar, Ashland, OR).
G-CSF administration
G-CSF (Amgen, Thousand Oaks, CA) was diluted in sterile phosphate buffered saline (PBS) and injected subcutaneously at 3.75 μg/100 μL every 12 hours.
In droplet immunostaining
Immunostaining of HSCs was adapted from a previously reported protocol.24 Glass slides with 4 mm diameter holes (#TF2404; Matsunami, Osaka, Japan) were coated with 20 μL 0.1% poly-L-lysine (#P8920; Millipore-Sigma), per dot, for 1 hour at room temperature. Slides were washed in distilled, deionized water, allowed to dry for 2 hours, 10 μL S-clone media (#1303; IWAI North America, San Carlos, CA) was added to each dot, and 150 HSCs were sorted directly into media using a slide-block adaptor. After sorting, the cells were allowed to rest for at least 1 hour at 4°C before 10 μL of 8% paraformaldehyde (PFA) was added to each droplet (final concentration, 4% PFA) and the cells were fixed for 20 minutes at room temperature. After washing with PBS, the cells were permeabilized with 20 μL of 0.1% Triton X-100 in PBS for 10 minutes at room temperature, blocked with 20 μL of block solution (5% goat serum in 0.1% Triton X-100) for 1 hour, and stained (refer to the supplemental Materials).
Statistical analysis
Data are presented as mean ± standard error of the mean (SEM) unless otherwise stated. Statistical significance was assessed using unpaired two-tailed Student t test or 2-way analysis of variance. GraphPad Prism (version 9.3.1) was used for all statistical analyses (GraphPad Software, La Jolla, CA).
Results
CD53 expression rapidly increases in HSCs during stress
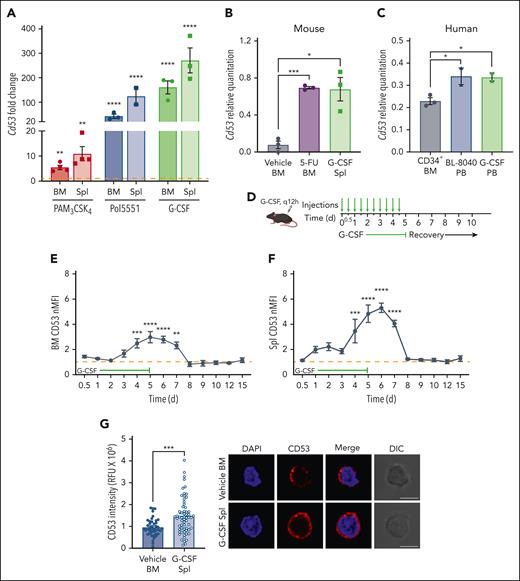
We performed microarray analyses of sorted HSCs (Lineage−, Sca-1+, c-kit+, CD48−, CD150+; gating strategy in supplemental Figure 1A) from WT mice treated with various inflammatory and proliferative stressors, including the Toll-like receptor 2 (TLR2) agonist PAM3CSK4, the CXCR4 antagonist Pol5551, and the cytokine G-CSF (Figure 1A). All treatments markedly increased Cd53 transcription above baseline, up to ∼200 fold higher, in both mobilized (spleen) HSCs and those that remained in the BM. Notably, Cd53 was one of the most highly upregulated genes in nearly all the treatment groups (Figure 1A; supplemental Tables 1-3). Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) confirmation of the microarray data showed that Cd53 expression in naïve HSCs (vehicle) was very low but was significantly upregulated by G-CSF stimulation or treatment with the chemotherapeutic agent 5-fluorouracil (5-FU; Figure 1B). Finally, CD34+ hematopoietic stem and progenitor cells (HSPCs) from the blood of healthy human donors treated with G-CSF or the CXCR4 antagonist BL-8040 upregulated CD53 compared with untreated CD34+ BM HSPCs (Figure 1C). Thus Cd53 is markedly upregulated in both mouse and human HSCs in response to various stressors.
CD53 expression rapidly increases in HSCs during stress. (A) Relative microarray expression values of Cd53 from sorted BM and spleen HSCs (KSL SLAM) from WT mice treated with the TLR2 agonist PAM3CSK4 (red; 25 μg injected intraperitoneally every 48 hours for 3 doses and analyzed 24 hours after the final dose), the CXCR4 antagonist Pol5551 (blue; 100 mg/kg per day for 2 weeks, delivered by osmotic pump), or the recombinant cytokine G-CSF (green; 3.75 μg injected subcutaneously every 12 hours for 7 days and analyzed 2 hours after the final dose) compared with vehicle-treated controls (orange dashed line). (B) Cd53 expression by qRT-PCR of RNA isolated from sorted HSCs after treatment with G-CSF (green; as above) or 5-FU (blue; 150 mg/kg injected intraperitoneally 6 days before analysis), compared with vehicle-treated controls. Relative quantitation of CD53 was calculated against actin using the 2−ΔCt method. (C) Human CD53 mRNA expression in G-CSF- and BL-8040-mobilized CD34+ cells compared with that in untreated CD34+ BM cells. Relative quantitation of CD53 was calculated against actin using 2−ΔCt method. (D) Outline of kinetic G-CSF injection and analysis experiment. WT mice were treated with 3.75 μg G-CSF every 12 hours, for up to 5 days and analyzed at designated time points. (E-F) Surface expression of CD53 from BM and splenic HSCs at the indicated times during treatment compared with vehicle-treated controls (orange dashed line; n = 3-5 mice per group). (G) Surface expression of CD53 (red) and DAPI (blue) assessed by immunofluorescence and confocal microscopy of WT HSCs from vehicle-treated BM and G-CSF-treated mobilized spleen HSCs (7 days of G-CSF; n = 2 mice per group with at least 20 individual HSCs imaged per mouse), with quantification shown on the left and a representative image of a single z-plane on the right. Error bars represent mean ± SEM. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001 by an unpaired Student t test. DAPI, 4',6-diamidino-2-phenylindole; nMFI, normalized median fluorescent intensity; PB, peripheral blood; Spl, spleen.
CD53 expression rapidly increases in HSCs during stress. (A) Relative microarray expression values of Cd53 from sorted BM and spleen HSCs (KSL SLAM) from WT mice treated with the TLR2 agonist PAM3CSK4 (red; 25 μg injected intraperitoneally every 48 hours for 3 doses and analyzed 24 hours after the final dose), the CXCR4 antagonist Pol5551 (blue; 100 mg/kg per day for 2 weeks, delivered by osmotic pump), or the recombinant cytokine G-CSF (green; 3.75 μg injected subcutaneously every 12 hours for 7 days and analyzed 2 hours after the final dose) compared with vehicle-treated controls (orange dashed line). (B) Cd53 expression by qRT-PCR of RNA isolated from sorted HSCs after treatment with G-CSF (green; as above) or 5-FU (blue; 150 mg/kg injected intraperitoneally 6 days before analysis), compared with vehicle-treated controls. Relative quantitation of CD53 was calculated against actin using the 2−ΔCt method. (C) Human CD53 mRNA expression in G-CSF- and BL-8040-mobilized CD34+ cells compared with that in untreated CD34+ BM cells. Relative quantitation of CD53 was calculated against actin using 2−ΔCt method. (D) Outline of kinetic G-CSF injection and analysis experiment. WT mice were treated with 3.75 μg G-CSF every 12 hours, for up to 5 days and analyzed at designated time points. (E-F) Surface expression of CD53 from BM and splenic HSCs at the indicated times during treatment compared with vehicle-treated controls (orange dashed line; n = 3-5 mice per group). (G) Surface expression of CD53 (red) and DAPI (blue) assessed by immunofluorescence and confocal microscopy of WT HSCs from vehicle-treated BM and G-CSF-treated mobilized spleen HSCs (7 days of G-CSF; n = 2 mice per group with at least 20 individual HSCs imaged per mouse), with quantification shown on the left and a representative image of a single z-plane on the right. Error bars represent mean ± SEM. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001 by an unpaired Student t test. DAPI, 4',6-diamidino-2-phenylindole; nMFI, normalized median fluorescent intensity; PB, peripheral blood; Spl, spleen.
To assess the kinetics of CD53 expression in HSCs, we measured the protein and mRNA levels of CD53 in WT mice treated with G-CSF for up to 5 days and allowed them to recover (Figure 1D). Cd53 RNA levels increased within a couple of hours of starting treatment and returned to baseline by 5 days after stopping (supplemental Figure 1B-C). In the first few days of treatment, CD53 surface expression was minimally upregulated (measured by normalized median fluorescent intensity; nMFI) compared with that in vehicle-treated controls (orange dashed line). However, after 4 days of G-CSF treatment, CD53 was significantly upregulated and sustained in both BM and spleen HSCs for up to 2 days after G-CSF cessation (Figure 1E-F). Notably, CD53 was upregulated by G-CSF in HSCs, but not in more differentiated progenitors (supplemental Figure 1D-E). CD53 upregulation was also observed by confocal microscopy of vehicle-treated BM HSCs compared with G-CSF mobilized splenic HSCs (Figure 1G). Thus, although CD53 is quickly upregulated in response to G-CSF treatment, its expression returns to baseline within days of stopping treatment, suggesting that CD53 functions acutely during HSC stress.
CD53 does not affect immunophenotypic HSC numbers or mobilization in response to G-CSF
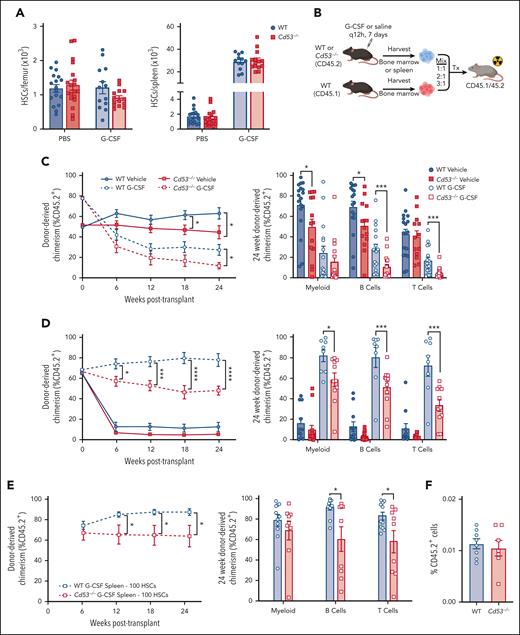
To investigate the role of CD53 in HSCs, we used our recently described Cd53-/- mouse model.16 Compared with WT littermates, Cd53−/− mice show no baseline differences in the frequency or absolute number of HSCs in the BM or spleen (Figure 2A; supplemental Figure 2A-B), with a modest increase in baseline circulating HSCs (supplemental Figure 2C). Furthermore, similar numbers of HSCs were mobilized in Cd53-/- mice compared with WT controls in response to G-CSF (Figure 2A; supplemental Figure 2B), suggesting that loss of CD53 does not impact baseline HSC development or mobilization. We noted modest decrease in BM HSC frequency after G-CSF (supplemental Figure 2A), although the absolute numbers were not significantly different from those in WT (Figure 2A). Finally, WT and Cd53−/− HSCs were equally quiescent at the baseline (supplemental Figure 2D).
Loss of CD53 does not affect HSC mobilization, but impairs HSC function in response to G-CSF. Mice were treated with 3.75 μg G-CSF every 12 hours for 7 days, with the final dose administered 2 hours before analysis. (A) Shown are absolute number of HSCs in the BM (left) and spleen (right) of vehicle- and G-CSF-treated Cd53−/− and WT mice (n = 13-19 mice per group; age, 6-10 weeks) over 6 independent experiments. (B) Outline of treated chimeric transplants. PBS-treated BM was transplanted at a ratio of 1:1, G-CSF-treated BM was transplanted at a ratio of 3:1, and both PBS- and G-CSF-treated spleens were transplanted at a ratio of 2:1, all competitively transplanted against the untreated whole BM. The donor mice were CD45.2, competitors were CD45.1, and the lethally irradiated recipients were CD45.1/45.2. Chimerism was assessed by peripheral blood flow cytometry analysis every 6 weeks. (C) Engraftment of whole BM from the indicated groups, including total leukocyte chimerism (left) and individual lineages at 24 weeks post-transplant (right). n = 13 to 18 recipients per group over 4 independent transplants, each with 1 donor (4 total donors). (D) Engraftment of whole splenocytes from the indicated groups, including total leukocyte chimerism (left) and individual lineages at 24 weeks post-transplant (right). n = 13 to 18 recipients per group over 4 independent transplants, each with one donor (4 total donors). (E) Engraftment of 100 sorted HSCs from the spleens of G-CSF treated WT or Cd53−/− mice, including total leukocyte chimerism (left) and individual lineages at 24 weeks post-transplant (right). n = 5 to 7 recipients per group over 2 independent transplants, with a total of 3 independent donors. (F) Marrow homing as assessed by transplanting sorted KSL (c-kit+, sca-1+, lineage-) cells from the spleens of G-CSF-treated WT and Cd53−/− mice into lethally irradiated CD45.1 WT mice, and enumerating CD45.2+ cells 20 hours later. n = 7 to 8 recipients in 2 independent transplants. Error bars represent mean ± SEM. ∗P < .05; ∗∗∗P < .001; and ∗∗∗∗P < .0001 by an unpaired Student t test.
Loss of CD53 does not affect HSC mobilization, but impairs HSC function in response to G-CSF. Mice were treated with 3.75 μg G-CSF every 12 hours for 7 days, with the final dose administered 2 hours before analysis. (A) Shown are absolute number of HSCs in the BM (left) and spleen (right) of vehicle- and G-CSF-treated Cd53−/− and WT mice (n = 13-19 mice per group; age, 6-10 weeks) over 6 independent experiments. (B) Outline of treated chimeric transplants. PBS-treated BM was transplanted at a ratio of 1:1, G-CSF-treated BM was transplanted at a ratio of 3:1, and both PBS- and G-CSF-treated spleens were transplanted at a ratio of 2:1, all competitively transplanted against the untreated whole BM. The donor mice were CD45.2, competitors were CD45.1, and the lethally irradiated recipients were CD45.1/45.2. Chimerism was assessed by peripheral blood flow cytometry analysis every 6 weeks. (C) Engraftment of whole BM from the indicated groups, including total leukocyte chimerism (left) and individual lineages at 24 weeks post-transplant (right). n = 13 to 18 recipients per group over 4 independent transplants, each with 1 donor (4 total donors). (D) Engraftment of whole splenocytes from the indicated groups, including total leukocyte chimerism (left) and individual lineages at 24 weeks post-transplant (right). n = 13 to 18 recipients per group over 4 independent transplants, each with one donor (4 total donors). (E) Engraftment of 100 sorted HSCs from the spleens of G-CSF treated WT or Cd53−/− mice, including total leukocyte chimerism (left) and individual lineages at 24 weeks post-transplant (right). n = 5 to 7 recipients per group over 2 independent transplants, with a total of 3 independent donors. (F) Marrow homing as assessed by transplanting sorted KSL (c-kit+, sca-1+, lineage-) cells from the spleens of G-CSF-treated WT and Cd53−/− mice into lethally irradiated CD45.1 WT mice, and enumerating CD45.2+ cells 20 hours later. n = 7 to 8 recipients in 2 independent transplants. Error bars represent mean ± SEM. ∗P < .05; ∗∗∗P < .001; and ∗∗∗∗P < .0001 by an unpaired Student t test.
Loss of CD53 impairs HSC repopulating activity
Next, we evaluated the HSC function using competitive transplants. WT and Cd53−/− mice (CD45.2) were treated with G-CSF or saline for 7 days. Whole BM (WBM) or spleen cells from treated mice were mixed with 1 × 106 WBM cells from the WT competitor (CD45.1) and transplanted into lethally irradiated WT recipients (CD45.1/45.2), and engraftment was monitored by peripheral blood chimerism (Figure 2B). Because G-CSF causes mobilization of HSCs to the spleen, both BM and spleen cells were transplanted into the donor mice.
In both vehicle- and G-CSF-treated BM recipients, loss of CD53 conferred a mild but significant reduction in multilineage repopulating activity (Figure 2C). More strikingly, splenic cells from G-CSF-treated Cd53−/− mice showed a marked decrease in multilineage repopulating activity compared with similarly treated cells from WT controls, with a reduction in engraftment from ∼80% to 50% (Figure 2D). A similar reduction in Cd53−/−HSC repopulating activity was noted when transplants were performed using 100 sorted HSCs from G-CSF-treated WT and Cd53−/− mice competitively transplanted with 2.5 × 105 WBM (Figure 2E). In addition to G-CSF, we also observed a reduction in the repopulating ability of Cd53−/− splenocytes after treatment with PAM3CSK4 (supplemental Figure 2E). Notably, BM homing was equivalent in Cd53−/− and WT HSPCs (Figure 2F).
RNA-sequencing identifies cycling dysregulation in Cd53−/− HSCs
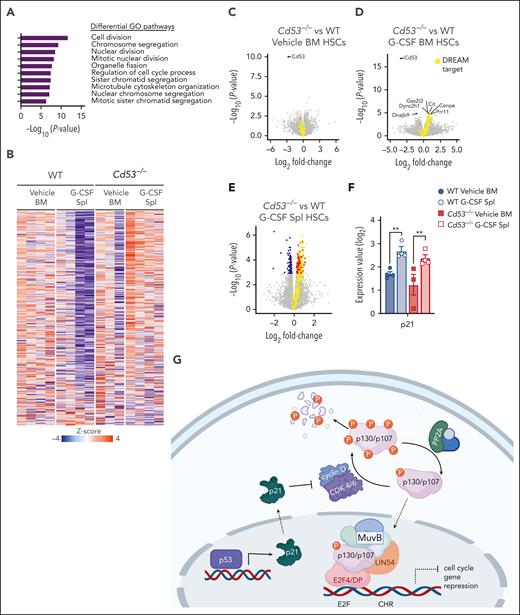
To investigate the mechanism by which CD53 protects HSC function in response to stress, we compared the transcriptomes of WT and Cd53−/− HSCs in the BM and spleen after G-CSF stimulation. Gene ontology (GO) pathway analysis revealed that the most significantly DEGs belonged to pathways regulating the cell cycle and division (Figure 3A). Notably, many of these genes belong to a group of ∼250 genes recently identified to be repressed by the transcriptional regulator DREAM complex.25 This complex is a transcriptional repressor induced by p53 and p21 in response to cellular stress, which subsequently downregulates the expression of genes involved in cell cycle and division. Nearly all DREAM complex target genes were repressed in G-CSF-treated WT splenic HSCs but upregulated in G-CSF-treated Cd53−/− splenic HSCs (Figure 3B). Comparing WT and Cd53−/− HSCs at baseline in the BM, the only significant DEG was Cd53 (Figure 3C). After 7 days of G-CSF, Cd53−/− HSCs that remained in the BM upregulated DREAM targets compared with WT Cenpe, Cit, Gas2I3, and Prr11; however, the most significant change was observed in the HSCs that had mobilized to the spleen (Figure 3D-E; supplemental Table 4). Consistent with their DREAM target gene repression, G-CSF mobilized HSCs have previously been shown to be quiescent,26 and GO pathway analysis of DEGs between WT BM and spleen HSCs following G-CSF in our study supports the finding that mobilized cells are more quiescent than those that remain in the marrow (supplemental Figure 3A). Notably, p21 (Cdkn1a) was equivalently induced by G-CSF treatment in both WT and Cd53−/− HSCs (Figure 3F), suggesting that the loss of DREAM activity in Cd53−/− HSCs is downstream of the initial sensing of cellular stress and production of p21. An overview of DREAM complex regulation is shown in Figure 3G.
RNA-sequencing identifies cycling dysregulation in Cd53−/− HSCs. (A) GO pathway analysis of DEGs (Padj < .05) comparing mobilized splenic HSCs from G-CSF-treated Cd53−/− and WT mice. (B) Heatmap of gene expression restricted to the known DREAM target genes in the indicated groups. (C-E) Volcano plots overlaying DREAM targets (yellow) over significantly downregulated (blue) and upregulated (red) genes between WT and Cd53−/− HSCs from vehicle-treated BM (C), G-CSF-treated BM (D), and G-CSF-treated spleens (E). The point corresponding to Cd53 was excluded from panel E to better show the differential expression pattern; a complete volcano plot can be seen in supplemental Figure 2F. (F) Expression of p21, a known regulator of DREAM complex formation, is equivalently upregulated by G-CSF treatment in both WT and Cd53−/− HSCs, as determined by bulk sequencing in panel A. (G) Overview of the DREAM complex regulation and function created using Biorender. Error bars represent mean ± SEM. ∗∗P < .01 by an unpaired Student t test.
RNA-sequencing identifies cycling dysregulation in Cd53−/− HSCs. (A) GO pathway analysis of DEGs (Padj < .05) comparing mobilized splenic HSCs from G-CSF-treated Cd53−/− and WT mice. (B) Heatmap of gene expression restricted to the known DREAM target genes in the indicated groups. (C-E) Volcano plots overlaying DREAM targets (yellow) over significantly downregulated (blue) and upregulated (red) genes between WT and Cd53−/− HSCs from vehicle-treated BM (C), G-CSF-treated BM (D), and G-CSF-treated spleens (E). The point corresponding to Cd53 was excluded from panel E to better show the differential expression pattern; a complete volcano plot can be seen in supplemental Figure 2F. (F) Expression of p21, a known regulator of DREAM complex formation, is equivalently upregulated by G-CSF treatment in both WT and Cd53−/− HSCs, as determined by bulk sequencing in panel A. (G) Overview of the DREAM complex regulation and function created using Biorender. Error bars represent mean ± SEM. ∗∗P < .01 by an unpaired Student t test.
Cd53−/− HSCs are less quiescent upon inflammatory stress
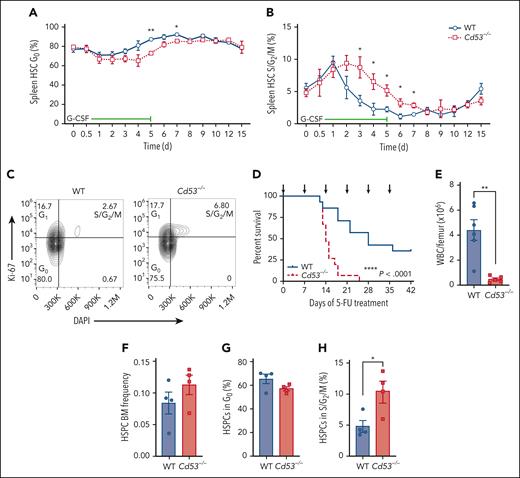
Next, we assessed the cycling status of mobilized HSCs using the same timed G-CSF treatment schedule as outlined in Figure 1D. The baseline cycling status was similar between WT and Cd53−/− HSCs, and they entered a cycle with similar kinetics (Figure 4A-B). However, the return to quiescence was delayed in the Cd53−/− HSCs (Figure 4A-B). An example of Ki-67/DAPI gating after 3 days of G-CSF treatment is shown (Figure 4C). There were no significant differences in the cycling status of BM HSCs between WT and Cd53−/− mice (supplemental Figure 3).
Cd53−/− HSCs are less quiescent upon inflammatory stress. Cell cycling in mobilized HSCs was measured in WT and Cd53−/− mice over the course of G-CSF treatment, as outlined in Figure 1D. The percentages of cells in the (A) G0 and (B) S/G2/M phases at the indicated time points are shown. (n = 3-6 mice per group per time point). (C) Representative flow plot of cell cycling analysis by Ki-67 and DAPI staining after 3 days of G-CSF treatment. (D) Survival of WT and Cd53−/− mice treated with serial 5-FU doses (arrows; n = 14-15 mice per group). (E) BM cellularity in WT and Cd53−/− mice at the time of death after serial 5-FU treatments. (n = 5-6 mice per group). (F) Frequency of HSPCs (lineage-, c-Kit+ cells) in WT and Cd53−/− BM at 7 days after a single dose of 5-FU. (G) Percentage of HSPCs in the G0 and (H) S/G2/M phases in the BM 7 days after a single dose of 5-FU. Error bars represent mean ± SEM. ∗P < .05; and ∗∗P < .01 by an unpaired Student t test. DAPI, 4',6-diamidino-2-phenylindole.
Cd53−/− HSCs are less quiescent upon inflammatory stress. Cell cycling in mobilized HSCs was measured in WT and Cd53−/− mice over the course of G-CSF treatment, as outlined in Figure 1D. The percentages of cells in the (A) G0 and (B) S/G2/M phases at the indicated time points are shown. (n = 3-6 mice per group per time point). (C) Representative flow plot of cell cycling analysis by Ki-67 and DAPI staining after 3 days of G-CSF treatment. (D) Survival of WT and Cd53−/− mice treated with serial 5-FU doses (arrows; n = 14-15 mice per group). (E) BM cellularity in WT and Cd53−/− mice at the time of death after serial 5-FU treatments. (n = 5-6 mice per group). (F) Frequency of HSPCs (lineage-, c-Kit+ cells) in WT and Cd53−/− BM at 7 days after a single dose of 5-FU. (G) Percentage of HSPCs in the G0 and (H) S/G2/M phases in the BM 7 days after a single dose of 5-FU. Error bars represent mean ± SEM. ∗P < .05; and ∗∗P < .01 by an unpaired Student t test. DAPI, 4',6-diamidino-2-phenylindole.
Consistent with prolonged cycling in Cd53−/− HSCs, Cd53−/− mice were less tolerant than their WT littermates to serial doses of 5-FU, which is toxic to the actively dividing cells. 5-FU was administered every 7 days, and Cd53−/− mice died significantly faster and had exacerbated BM hypocellularity (Figure 4D-E). Assessment of BM HSPC (Lineage− c-Kit+) cycling status confirmed enhanced cycling of Cd53−/− cells 7 days after a single dose of 5-FU (Figure 4F-H). Furthermore, transcriptome analysis of HSCs 10 days after a dose of 5-FU revealed enhanced DREAM target expression by gene set enrichment analysis in Cd53−/− HSCs compared with WT (supplemental Figure 4; supplemental Table 5). Taken together, these data suggest that CD53 promotes the return of HSCs to quiescence after stress-induced cycling, with the loss of CD53 resulting in prolonged cycling and reduced HSC function.
Inhibition of cell cycling with palbociclib equalizes the function of WT and Cd53−/− HSCs
To confirm that the loss of CD53 primarily affects HSC function through cycling dysregulation, mice were treated with the cyclin-dependent kinase (CDK) 4/6 inhibitor, palbociclib. One key step in the regulation of the DREAM complex is the phosphorylation of the pocket protein family members p107 (retinoblastoma like 1; Rbl1) and p130 (Rbl2) by CDK4/6–cyclin D dimers.27 These proteins are phosphorylated during the G1 checkpoint and S-phase transition, rendering them unable to bind E2F4 and inhibit cell cycle progression. Phosphorylated p107 (p-p107) and p-p130 can then be ubiquitinated and targeted for degradation28 or dephosphorylated and recycled.29 By inhibiting the phosphorylation of p107 or p130, palbociclib promotes DREAM complex activity and inhibits cycling.
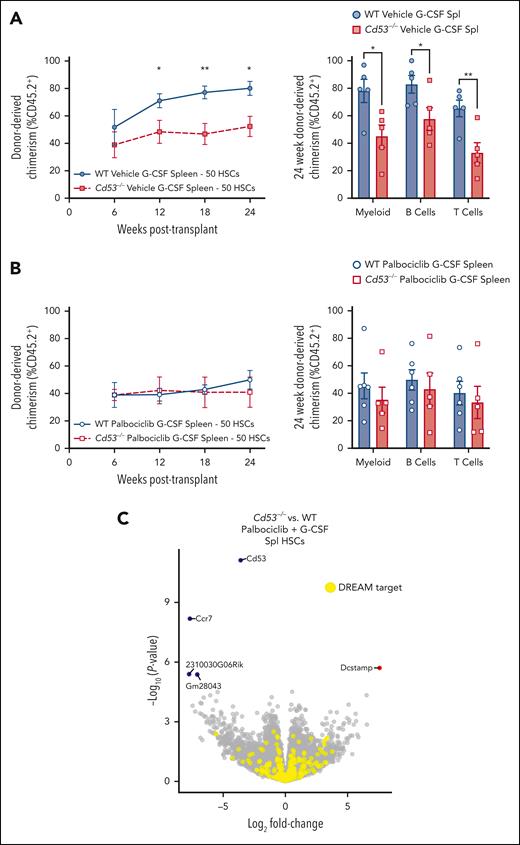
Mice were treated with palbociclib or vehicle control, beginning 1 day before 7 days of G-CSF administration. Then, 50 splenic mobilized HSCs were sorted and competitively transplanted against 2.5 × 105 WBM into lethally irradiated recipients. Both WT and Cd53−/− vehicle-treated G-CSF HSCs performed similarly to prior transplants (Figures 2D-E) with Cd53−/− HSCs engrafting worse than WT (Figure 5A). However, when treated with palbociclib, G-CSF mobilized HSCs from WT and Cd53−/− mice performed equivalently to each other (Figure 5B). Furthermore, transcriptome comparison between palbociclib- G-CSF-treated mobilized HSCs from WT and Cd53−/− mice revealed only 5 significant DEGs, none of which were targets of the DREAM complex (Figure 5C; supplemental Table 6). In addition to equalizing HSC function, inhibition of cycling with palbociclib negated nearly all DEGs in G-CSF-treated HSCs between WT and Cd53−/− mice, suggesting that the function of CD53 in HSCs acts largely or exclusively through cell cycle regulation.
Inhibition of cell cycling with palbociclib equalizes the function of WT and Cd53−/− HSCs. WT and Cd53−/− mice were treated with palbociclib or vehicle control, daily via oral gavage, beginning 1 day before 7 days of G-CSF treatment, and 50 sorted HSCs were competitively transplanted against 2.5 × 105 WBM into lethally irradiated recipients. (A) Engraftment of vehicle controls (no palbociclib), including total leukocyte chimerism (left) and individual lineages at 24 weeks post-transplant (right). n = 5 recipients per group in 2 independent experiments. (B) Engraftment of palbociclib treated groups, including total leukocyte chimerism (left) and individual lineages at 24 weeks post-transplant (right). n = 5 to 6 recipients per group in 2 independent experiments. (C) Volcano plot of DEGs (Padj < .05) comparing mobilized splenic HSCs from WT and Cd53−/− mice treated with G-CSF and palbociclib. Error bars represent mean ± SEM. ∗P < .05; and ∗∗P < .01 by an unpaired Student t test.
Inhibition of cell cycling with palbociclib equalizes the function of WT and Cd53−/− HSCs. WT and Cd53−/− mice were treated with palbociclib or vehicle control, daily via oral gavage, beginning 1 day before 7 days of G-CSF treatment, and 50 sorted HSCs were competitively transplanted against 2.5 × 105 WBM into lethally irradiated recipients. (A) Engraftment of vehicle controls (no palbociclib), including total leukocyte chimerism (left) and individual lineages at 24 weeks post-transplant (right). n = 5 recipients per group in 2 independent experiments. (B) Engraftment of palbociclib treated groups, including total leukocyte chimerism (left) and individual lineages at 24 weeks post-transplant (right). n = 5 to 6 recipients per group in 2 independent experiments. (C) Volcano plot of DEGs (Padj < .05) comparing mobilized splenic HSCs from WT and Cd53−/− mice treated with G-CSF and palbociclib. Error bars represent mean ± SEM. ∗P < .05; and ∗∗P < .01 by an unpaired Student t test.
CD53 supports p107 and p130 protein expression
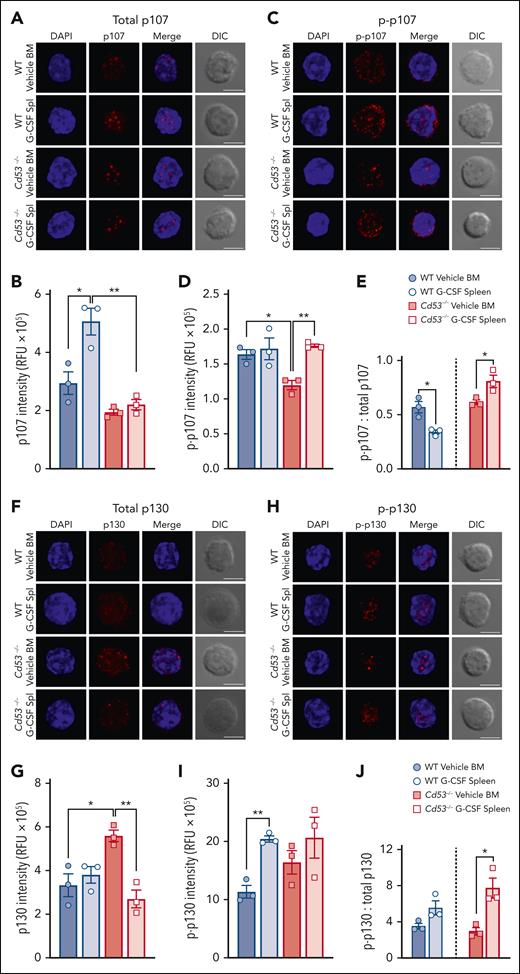
Tetraspanins affect cellular functions via the organization of protein interaction networks at the cell membrane, where they influence the stabilization, modification, and/or function of their interacting partners. Thus, we determined whether the loss of CD53 affected the protein levels of the DREAM complex components. HSCs from WT and Cd53−/− mice treated with vehicle or G-CSF were stained for the immunofluorescence of p107 and p130. Individual HSCs were imaged using confocal microscopy, and protein expression was measured using relative fluorescence units (RFU). Total protein levels of p107 increased in WT, but not in Cd53−/− HSCs, after G-CSF treatment (Figure 6A-B; supplemental Figure 5A). Expression of Thr369 p-p107 was equivalent after G-CSF treatment in WT HSCs, but increased in Cd53−/− HSCs with G-CSF (Figure 6C-D; supplemental Figure 5B). Because the function of pocket proteins is highly dependent on their phosphorylation status, we calculated the ratio of p-p107 to total p107 in these samples. Hypophosphorylation of p107 and p130 is associated with increased DREAM complex binding and gene repression, whereas hyperphosphorylation targets these proteins for degradation. The ratio of p-p107 to total p107 decreased with G-CSF in WT HSCs but increased in Cd53−/− HSCs, suggesting reduced p107 availability for DREAM complex engagement in Cd53−/− HSCs (Figure 6E).
CD53 supports p107 and p130 protein expression. (A) Representative maximum projection confocal images of total p107 (red) and DAPI (blue) in individual HSCs with a single XY differential interference contrast (DIC) image. (B) Quantification of the total p107 intensity in the RFU from HSCs from the indicated groups (n = 30-50 cells per group) over 3 independent experiments. Average RFU per mouse from at least 20 individual HSCs per sample is shown. The average spread in a single mouse can be seen in supplemental Figure 5A. (C) Representative confocal images of Thr369 phospho-p107 (red) and DAPI (blue) in individual HSCs. (D) Quantification of Thr369 p-p107 intensity in HSCs from the indicated groups (n = 29-42 cells per group) in 3 independent experiments. The average RFU per mouse from at least 20 individual HSCs per sample is shown. The average spread in a single mouse can be seen in supplemental Figure 5B. (E) The ratio of p-p107 to total p107. (F) Representative confocal images of total p130 (red) and DAPI (blue) in individual HSCs. (G) Quantification of total p130 in RFU from HSCs from the indicated groups (n = 37-66 cells per group) in 3 independent experiments. The average RFU per mouse from at least 20 individual HSCs per sample is shown. The average spread in a single mouse can be seen in supplemental Figure 5C. (H) Representative confocal images of Ser639 p-p130 (red) and DAPI (blue) in individual HSCs. (I) Quantification of Ser639 p-p130 intensity in HSCs from the indicated groups (n = 35-41 cells per group) over 3 independent experiments. The average RFU per mouse from at least 20 individual HSCs per sample is shown. The average spread in a single mouse can be seen in supplemental Figure 5D. (J) The ratio of p-p130 to total p130. All fluorescent images are maximum intensity projections, whereas the DIC images are of a single XY plane. All scale bars are 5 μm. Error bars represent the mean ± standard deviation. ∗P < .05; and ∗∗P < .01 by an unpaired Student t test. DAPI, 4',6-diamidino-2-phenylindole.
CD53 supports p107 and p130 protein expression. (A) Representative maximum projection confocal images of total p107 (red) and DAPI (blue) in individual HSCs with a single XY differential interference contrast (DIC) image. (B) Quantification of the total p107 intensity in the RFU from HSCs from the indicated groups (n = 30-50 cells per group) over 3 independent experiments. Average RFU per mouse from at least 20 individual HSCs per sample is shown. The average spread in a single mouse can be seen in supplemental Figure 5A. (C) Representative confocal images of Thr369 phospho-p107 (red) and DAPI (blue) in individual HSCs. (D) Quantification of Thr369 p-p107 intensity in HSCs from the indicated groups (n = 29-42 cells per group) in 3 independent experiments. The average RFU per mouse from at least 20 individual HSCs per sample is shown. The average spread in a single mouse can be seen in supplemental Figure 5B. (E) The ratio of p-p107 to total p107. (F) Representative confocal images of total p130 (red) and DAPI (blue) in individual HSCs. (G) Quantification of total p130 in RFU from HSCs from the indicated groups (n = 37-66 cells per group) in 3 independent experiments. The average RFU per mouse from at least 20 individual HSCs per sample is shown. The average spread in a single mouse can be seen in supplemental Figure 5C. (H) Representative confocal images of Ser639 p-p130 (red) and DAPI (blue) in individual HSCs. (I) Quantification of Ser639 p-p130 intensity in HSCs from the indicated groups (n = 35-41 cells per group) over 3 independent experiments. The average RFU per mouse from at least 20 individual HSCs per sample is shown. The average spread in a single mouse can be seen in supplemental Figure 5D. (J) The ratio of p-p130 to total p130. All fluorescent images are maximum intensity projections, whereas the DIC images are of a single XY plane. All scale bars are 5 μm. Error bars represent the mean ± standard deviation. ∗P < .05; and ∗∗P < .01 by an unpaired Student t test. DAPI, 4',6-diamidino-2-phenylindole.
Comparing the total levels of p130 protein, WT HSCs had equivalent expression between vehicle- and G-CSF-treated samples; however, Cd53−/− HSCs had heightened baseline levels compared with WT HSCs and significantly reduced p130 expression upon G-CSF treatment compared with vehicle-treated Cd53−/− HSCs (Figure 6F-G; supplemental Figure 5C). Quantification of Ser639 p-p130 in HSCs revealed significantly increased phosphorylation of WT HSCs with G-CSF, but no significant difference was observed in Cd53−/− HSCs (Figure 6H-I; supplemental Figure 5D). In WT HSCs, the ratio of p-p130 to total p130 was nearly equal upon G-CSF treatment. However, in Cd53−/− HSCs, the ratio of p-p130 to total p130 increased after G-CSF treatment, again suggesting reduced p130 availability for DREAM complex engagement (Figure 6J). Notably, the transcript levels of both p107 and p130 were similar in the WT and Cd53−/− HSCs (supplemental Table 4). Together, these data suggest that CD53 regulates p107 and p130 protein levels, promoting the availability of active hypophosphorylated p107/p130 for the DREAM complex activity.
CD53 promotes p130-mediated DREAM complex binding
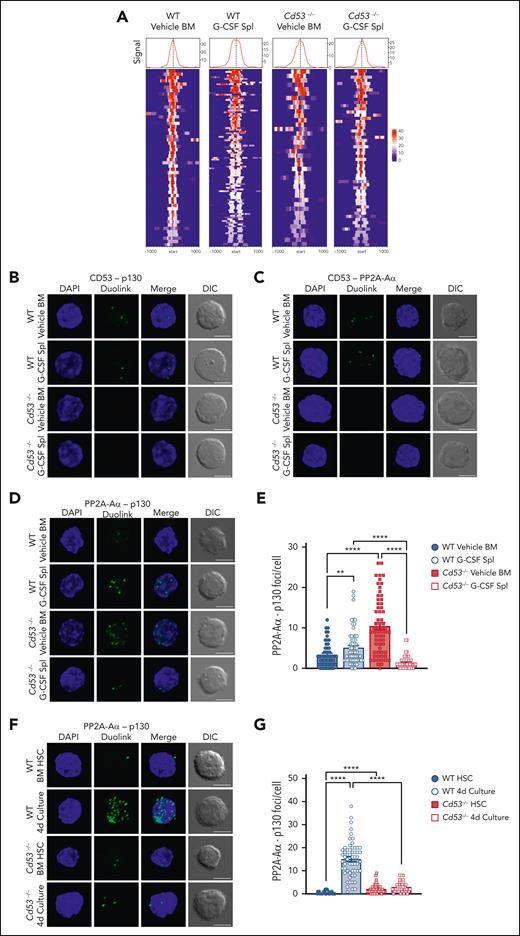
Our data predicted that the loss of CD53 would reduce the DREAM complex activity. To test this, we performed CUT&Tag sequencing in WT and Cd53−/− HSCs under naïve (vehicle-treated BM) and stressed (G-CSF-treated spleen) conditions to measure p130 binding to DREAM target genes. Library preparations from 50 000 HSPCs (KSL CD48−) were performed with an anti-p130 antibody using the CUT&Tag protocol developed by the Henikoff lab.30 Peak counts around DREAM target genes were highest in the G-CSF mobilized WT cells (Figure 7A; supplemental Table 7), consistent with the observation that DREAM target gene expression was reduced in G-CSF-treated WT HSCs, but not in Cd53−/− HSCs (Figure 3B).
CD53 promotes DREAM complex binding through association with p130 and its phosphatase PP2A. (A) CUT&Tag sequencing of WT and Cd53−/− naïve BM- and G-CSF-treated splenic HSPCs. p130 read intensities within 100 kb of the transcription start site of the DREAM targets are shown. See supplemental Table 7 for peak counts and DREAM enrichment. The Duolink proximity ligation assay reveals an interaction between (B) CD53-p130 and (C) CD53-PP2A-Aα in WT HSCs, with no signal in Cd53−/− HSC controls. Representative maximum intensity projection confocal images of Duolink foci (green) and DAPI (blue) are shown, indicating protein-protein interaction within 40 nm, with a single XY DIC image. (D) Representative maximum intensity projection confocal images of the PP2A-Aα-p130 Duolink interaction (green) in WT and Cd53−/− HSCs at baseline and after G-CSF treatment. Loss of CD53 impairs the interaction between p130 and the PP2A phosphatase structural subunit, PP2A-Aα, (E) quantified as the number of foci per HSC (n = 55-63 cells per group) in 2 independent experiments. (F) Representative maximum intensity projection confocal images of the PP2A-Aα-p130 Duolink interaction (green) in naïve HSCs and HSCs after 4 days of culture in StemSpan, with 100 ng/mL TPO and 100 ng/mL SCF, and (G) quantified as the number of foci per HSC (n = 42-57 cells per group) in 2 independent experiments. Notably, although the number of foci is not an exact count of the total interactions per cell, it provides a relative measure between groups. Error bars represent mean ± SEM. All scale bars are 5 μm. ∗∗P < .01; and ∗∗∗∗P < .0001 by an unpaired Student t test. DAPI, 4',6-diamidino-2-phenylindole.
CD53 promotes DREAM complex binding through association with p130 and its phosphatase PP2A. (A) CUT&Tag sequencing of WT and Cd53−/− naïve BM- and G-CSF-treated splenic HSPCs. p130 read intensities within 100 kb of the transcription start site of the DREAM targets are shown. See supplemental Table 7 for peak counts and DREAM enrichment. The Duolink proximity ligation assay reveals an interaction between (B) CD53-p130 and (C) CD53-PP2A-Aα in WT HSCs, with no signal in Cd53−/− HSC controls. Representative maximum intensity projection confocal images of Duolink foci (green) and DAPI (blue) are shown, indicating protein-protein interaction within 40 nm, with a single XY DIC image. (D) Representative maximum intensity projection confocal images of the PP2A-Aα-p130 Duolink interaction (green) in WT and Cd53−/− HSCs at baseline and after G-CSF treatment. Loss of CD53 impairs the interaction between p130 and the PP2A phosphatase structural subunit, PP2A-Aα, (E) quantified as the number of foci per HSC (n = 55-63 cells per group) in 2 independent experiments. (F) Representative maximum intensity projection confocal images of the PP2A-Aα-p130 Duolink interaction (green) in naïve HSCs and HSCs after 4 days of culture in StemSpan, with 100 ng/mL TPO and 100 ng/mL SCF, and (G) quantified as the number of foci per HSC (n = 42-57 cells per group) in 2 independent experiments. Notably, although the number of foci is not an exact count of the total interactions per cell, it provides a relative measure between groups. Error bars represent mean ± SEM. All scale bars are 5 μm. ∗∗P < .01; and ∗∗∗∗P < .0001 by an unpaired Student t test. DAPI, 4',6-diamidino-2-phenylindole.
CD53 interacts with p130 and its phosphatase PP2A
The exact mechanism by which plasma membrane-bound tetraspanin CD53 affects pocket protein levels and DREAM complex binding to DNA was not explicitly obvious. Both p107 and p130 are regulated by the phosphorylation of CDK4/6–cyclin D dimers and by dephosphorylation via the serine/threonine PP2A.29 The latter allows pocket protein recycling, prevents ubiquitin-mediated degradation, and promotes their nuclear translocation,31 thereby stimulating participation in DREAM complex binding. Thus, we hypothesized that CD53 interacts with the pocket proteins and PP2A to regulate the DREAM complex. Notably, these proteins shuttle between the nucleus and cytoplasm, and thus could interact near the membrane where CD53 is primarily localized (Figure 1G).32 We particularly focused on p130, as its total levels dropped significantly with cytokine stress in the absence of CD53 (Figure 6G).
To determine these interactions, we utilized the Duolink proximity ligation assay (PLA) system, which can determine protein-protein interactions within 40 nm of each other through oligonucleotide-conjugated antibodies and rolling circle amplification. HSCs from vehicle- and G-CSF-treated WT and Cd53−/− mice were sorted onto slides and Duolink chemistry was performed. By applying primary antibodies to CD53 and p130, Duolink foci were detected in both vehicle- and G-CSF-treated WT HSCs, indicating that CD53 and p130 interacted with each another, and all Duolink signals were lost in the absence of CD53 (Figure 7B; supplemental Figure 5E). A similar investigation of CD53 and the PP2A-Aα subunit of PP2A confirmed the interaction between these proteins (Figure 7C; supplemental Figure 5F), which is consistent with a recent report that identified PP2A as a CD53 binding partner.33 The interaction of CD53 with p130 was confirmed by the expression of FLAG-tagged CD53 in HEK293T cells and the performance of reciprocal co-immunoprecipitation experiments (supplemental Figure 5H-I).
We next reasoned that, because CD53 interacts individually with both p130 and its phosphatase, it may facilitate the interaction between these 2 proteins. To test this, we performed PLA to measure the amount of PP2A-Aα– p130 interaction in WT and Cd53−/− HSCs. Rather than confirming protein-protein interactions, as these proteins are known to interact, the Duolink assay provides a quantitative measure of this interaction. In WT HSCs, we observed a modest but significant increase in the number of PP2A-Aα– p130 foci per cell from vehicle- to G-CSF-treated cells (Figure 7D; supplemental Figure 5G). However, in Cd53−/− HSCs, although the number of PP2A-Aα– p130 foci at baseline was higher than that in WT HSCs, these interactions were almost completely lost upon G-CSF treatment (Figure 7D-E).
In addition to G-CSF treatment, naïve HSCs from WT and Cd53−/− mice were cultured ex vivo for 4 days and stained for PP2A-AΑ–p130 interaction. Similar to G-CSF treated HSCs, cultured WT cells showed significant increase in the interaction between PP2A-Aα and p130, in a CD53-dependent manner (Figure 7F-G). Finally, treatment with PAM3CSK4 also led to a CD53- dependent trend toward increase in the PP2A-Aα-p130 interaction (supplemental Figure 6). Thus, multiple stressors promoted CD53-dependent interactions between p130 and the PP2A in HSCs. Together, these data suggest that CD53 interacts with both p130 and PP2A subunit PP2A-Aα in response to stress, promoting their interaction and increasing the amount of p130 available for DREAM complex binding. This was consistent with the finding of relatively high levels of phosphorylated p130 and reduced total protein levels in the absence of CD53.
Discussion
Quiescence is a cardinal feature of HSCs, and the most robust long term repopulating activity is associated with quiescent populations of HSCs. Nevertheless, HSCs must respond to inflammatory and proliferative cues, allowing the production of effector cells, as needed, to fight infection and other insults. After a brief increase in cycling, HSCs must return to quiescence to avoid proliferative exhaustion and maintain their functional integrity.1,11 The ability to limit their proliferative activity in response to stress is thus a crucial attribute of HSCs. However, the braking mechanisms that allow for dynamic variation in HSC cycling are not well understood. In this study, we show that tetraspanin CD53 is upregulated in mouse and human HSCs in response to a variety of inflammatory and proliferative stressors, and our data support a model whereby this upregulation of CD53 facilitates the return of cycling HSCs to quiescence via the promotion of DREAM complex activity.
CD53 was previously identified as one of the few proteins that segregate differentially in dividing human HSCs, localizing to a more functionally primitive population.20 However, the specific role of CD53 in the regulation of HSCs has not been previously described. Notably, other tetraspanins have been shown to regulate HSC quiescence, including CD81, CD82, and CD63. CD81 was reported to promote HSC quiescence after 5-FU-stimulated proliferation via deactivation of the protein kinase B (Akt) pathway.34 CD82 was found to regulate HSC BM homing,35 and to facilitate the interaction between HSCs and macrophages in the BM to promote quiescence via TGFβ signaling.36 Similarly, CD63 was recently reported to regulate TGFβ signaling and quiescence in HSCs.37 Thus, CD53 is among a growing list of tetraspanin family members that influence HSC cycling, however the mechanism by which it does so uniquely involves interaction with p130 and regulation of DREAM complex activity. This mechanism is supported by the fact that enforced DREAM activity and suppression of cell cycling by palbociclib equalized the functions of CD53 and WT HSCs. Moreover, palbociclib treatment removed nearly all DEGs between G-CSF-treated Cd53−/− and WT HSCs, suggesting that cell cycle control may be the primary function of CD53 in stressed HSCs.
Previous studies have shown that DREAM activity is necessary for processes such as embryonic development, tumor suppression, and chemotherapy-induced apoptosis,38,39 however the role of the DREAM complex in the HSC response to inflammatory stress has not been previously described.25 Under homeostatic conditions, loss of p130 confers no obvious blood phenotype,40 and loss of p107 results in mild myeloid hyperplasia that is only evident in a Balb/c genetic background.41 Simultaneous loss of all 3 Rb family members (Rb, p107, and p130) in adult HSCs, however, leads to hyperproliferation of HSCs, apoptosis of lymphoid progenitors, and the development of a lethal myeloproliferative disease in mice.42 Thus, Rb family members are important regulators of HSC quiescence and the balance between lymphoid and myeloid cell production, and these proteins can largely compensate for one another in the absence of cellular stress (given the relatively mild phenotypes of individual knockouts). However, the specific roles of the DREAM-related pocket proteins p107 and p130, in inflammatory or proliferative stress have not been described. Our data showed that the relative amounts of hypophosphorylated p107 and p130 increased with stress, consistent with enhanced DREAM activity and repression of cycling-associated genes. However, in the absence of CD53, stressed HSCs downregulate p107 and p130 protein levels, with an increased relative amount of phosphorylated (inactive) protein, and our Duolink studies suggest that CD53 facilitates the interaction of p130 with its primary phosphatase, PP2A-Aα. Notably, the cycling status and DREAM target expression in naïve BM HSCs were not significantly altered by loss of CD53. In fact, the total p130 protein levels were increased, suggesting that the role of CD53 in DREAM regulation and HSC cycling suppression is specific to the stressed state, consistent with the fact that the expression of CD53 is low in the absence of stress.
Taken together, our data suggest a model whereby CD53 promotes the activity of pocket proteins in response to HSC stress, facilitating DREAM complex binding and returning to quiescence. Alternatively, CD53 may influence DREAM activity via additional binding interactions that were not specifically tested in the present study. Further studies are needed to more comprehensively assess the interacting partners of CD53 in HSCs and determine the nature- direct vs indirect- of these interactions. In addition, although CD53 appears to be primarily expressed on the cell surface of stressed HSCs, further studies are needed to more definitively elucidate the subcellular localization of its interactions with DREAM-associated proteins.
Finally, a differential response to inflammatory stress contributes to the expansion of mutant HSCs in aging- related clonal hematopoiesis (CH).43 CH involves the clonal expansion of HSCs with leukemia-associated mutations, most commonly in TET2 or DNMT3A, and confers an increased risk of hematopoietic malignancy, cardiovascular disease, and all-cause mortality.44 HSCs bearing mutations in Tet2 or Dnmt3a expand relative to their normal counterparts in response to infection or exposure to inflammatory cytokines, suggesting that they are comparatively resistant to replicative exhaustion.7,45,46 The reasons for their differential response to inflammatory signals are not entirely clear. Interestingly, HSCs lacking Tet2 or Dnmt3a express significantly higher levels of CD53 than WT HSCs,47 and it is therefore tempting to speculate that CD53 may help facilitate their clonal advantage and resistance to replicative stress. Thus, future studies are needed to determine whether CD53 protects mutant HSCs against inflammatory and proliferative stress in a similar fashion to WT HSCs, and whether chronic upregulation of CD53 is beneficial to HSC function.
Acknowledgments
The authors thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital for the use of the Siteman Flow Cytometry Core. They thank Jackie Tucker-Davis and Patrick Shumway for animal care. They the Genome Engineering and iPSC Center at Washington University for the of generation of the Cd53−/− mice.
This work was supported by funding from The Children’s Discovery Institute of St. Louis Children’s Hospital Foundation (C.J.L. and L.G.S.), the National Heart, Lung, and Blood Institute (R01 HL134896; L.G.S.), the National Institute of Diabetes and Digestive and Kidney Diseases (R01DK114047; C.J.L.) and the Training Program in Cellular and Molecular Biology (T32 GM007067-44; Z.J.G.)
Authorship
Contribution: Z.J.G and L.G.S designed the research; Z.J.G., D.A.M., Q.D., W.L., N.R., R.G., L.C.P., and W.Y. performed and analyzed the results of the experiments; Z.J.G. and W.Y. prepared the figures; M.R. and C.J.L provided critical resources; Z.J.G. and L.G.S. with the inputs of C.J.L. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Laura G. Schuettpelz, Department of Pediatrics, Washington University School of Medicine, 660 S. Euclid Ave, St. Louis, MO 63110; e-mail: schuettpelz_l@wustl.edu.
References
Author notes
RNA-seq data reported in this article have been deposited in the Gene Expression Omnibus (GEO) database (accession number GSE219050). More details of materials and methods are provided in the supplemental Data.
Data are available on request from the corresponding author, Laura G. Schuettpelz (schuettpelz_l@wustl.edu).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal