Abstract
Multiple myeloma (MM) is a heterogenous disease in which clonal evolution or preexisting reservoirs of phenotypically divergent cells may be harbingers of therapeutic resistance. Prior research has suggested the presence of myeloma cells with immunophenotypic and expression profiles dedifferentiated from that of terminally differentiated plasma cells that may be found in circulation, marrow, or potentially other lymphoid compartments; however, this has not been conclusively demonstrated in patients. The identification of such clonotypic myeloma cells could lead to a better understanding of myeloma biology and resistance, and inform therapies targeted at these populations, and single-cell technologies provide the granularity to characterize them. Next-generation sequencing (NGS)-based measurable residual disease (MRD) clinical-grade assays (eg. clonoSEQ®, Adaptive Biosciences) identify individuals’ unique heavy and light chain sequences. Theoretically this sequencing data could be overlaid with research-grade single-cell multiomics to identify clusters of clonotypic cells with variable differentiation. In this pilot study using the Enhanced Single Cell Analysis with Protein Expression (ESCAPE) platform (Singleron Biotechnologies) we aimed to identify peripherally circulating clonotypic MM cells by their NGS-MRD sequence and simultaneously determine whether their multiomic profile reflected possible dedifferentiated clusters found in paired marrow.
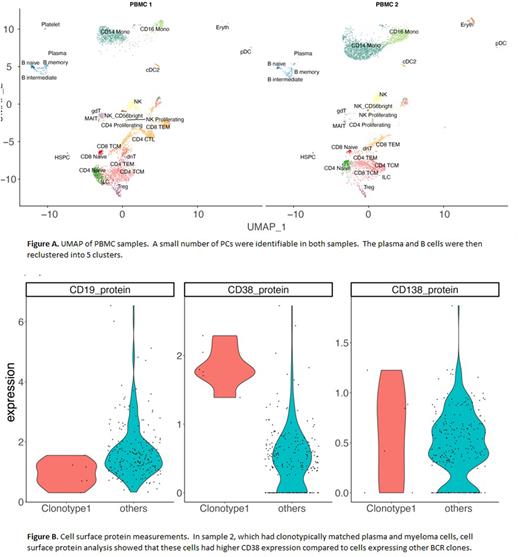
Using scRNA-Seq (10x Genomics) data previously generated at our site from bone marrow mononuclear cells we identified MM cell clusters enriched for differentially expressed genes known to be expressed in B-cells and plasmablasts. These genes included CD19, PAX5, BCL6, EZH2, STMN1, HMGB2, MKI67 and CD27. We next sought to assay the peripheral blood mononuclear cells (PBMC) of patients (n=2) with an overrepresentation of cells in these clusters. To evaluate both the transcriptomic and immunophenotypic profiles of the PBMCs at single-cell resolution we utilized Singleron's Immune Profiling panel on the ESCAPE platform. Using Singleron's MapSuite™ analysis pipeline, we annotated cell-type classifications, and highlighted cells within the mature B-cell/plasma cell differentiation spectrum (Figure A). Within this subset, we attempted to identify gene expression reflective of the previously identified "dedifferentiated” marrow clusters with specific genes of interest, but found that there was not significant overlap. Heatmaps comparing the 5 peripheral subclusters with marrow plasma cells demonstrate some concordance with the marrow subclusters, with examination of these overlaps ongoing, but these were not the "dedifferentiated” marrow clusters.
The included patients also had clonoSEQ® identification sequences of heavy chain and/or light chain available. By cross-referencing the NGS-MRD sequence data (clonoSEQ®, Adaptive Biosciences), we were able to use ESCAPE-Seq to identify definitive clonotypic circulating MM cells within the PBMC samples at single cell granularity. Among PBMCs from sample 2 we were able to identify circulating clonotypic myeloma cells with 100% match to the IgH and IgK MRD sequences, which also represented the dominant V(D)J clonotype in peripheral circulating. Concomitant single-cell surface protein analysis of this clonotypic cell population demonstrated significant expression of CD38, without significant differential expression of CD138 nor surface markers of B-cells (Figure B).
While these data are preliminary and studies are ongoing, we successfully leveraged clinical-level MRD sequencing data to inform single-cell multiomics, a potentially powerful translational research pathway for any hematologic malignancy in which NGS-MRD is clinically available. We definitively identified a small subset of circulating MM cells and used the ESCAPE platform to efficiently generate a multiomics profile. This profile suggested elevated CD38 expression without elevated CD138 expression, which previously has been associated with dedifferentiated plasma cells and plasmablasts, although further analyses are ongoing. In future studies, we will enrich for B-lymphoid spectrum cells prior to single-cell analytics.
Disclosures
Goldsmith:Wugen Inc.: Consultancy; Sanofi-Genzyme: Consultancy; Janssen: Consultancy. Jayasinghe:Wugen: Consultancy; Multiple Myeloma Research Foundation: Consultancy. Vij:BMS: Honoraria, Other: Grant support; Takeda: Honoraria, Other: Grant support; Beigene: Honoraria; adaptive: Honoraria; oncopetideslegend: Honoraria; Janssen: Honoraria; GSK: Honoraria; Sanofi: Honoraria, Other: Grant support.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal