Abstract
Introduction Alterations in IKZF1, including the IKZF1plusgenetic profile, have been associated with poor prognosis in patients with B-acute lymphoblastic leukemia (B-ALL). Although the types and frequencies of IKZF1 alterations vary by leukemia subtype, the effects of IKZF1 alteration status and leukemia subtypes on clinical outcome are not fully understood. We have assessed the spectrum of IKZF1 alterations, including IKZF1plus, IKZF1 deletions only, the IKZF1 IK6 dominant negative mutant, sequence mutations, and their associations with clinical outcome, in the context of genomic subtypes of B-ALL.
Patients and Methods We analyzed single nucleotide polymorphism 6.0 microarrays, whole genome sequencing, and whole exome sequencing data to detect genetic alterations in a cohort of 688 pediatric patients with B-ALL enrolled into St. Jude Total Therapy XV and XVI studies. The IKZF1plus genetic profile was denoted by the presence of IKZF1 deletions together with one or more of the following alterations: deletion in PAX5, CDKN2A, or both CDKN2B loci, or CRLF2-rearrangement, in the absence of copy number alterations in ERG or DUX4-rearrangement.
Results Alterations in IKZF1 were detected in 11.8% of patients (81 out of 688), with IKZF1 deletions, sequence mutations, and concurrent deletion and sequence mutations present in 8.1%, 3.2%, and 0.4% of patients, respectively. The IKZF1plus profile was detected in 4.2% of patients and IKZF1 deletions only detected in 3.9% of patients. Of 29 patients with IKZF1plus, 51.7% had unfavorable subtypes of leukemia (including Ph and Ph-like-CRLF2), 27.6% had intermediate subtypes of leukemia, and 12.8% had favorable subtypes (DUX4, ETV6-RUNX1, and Hyperdiploid) of leukemia compared to patients with IKZF1 deletions only, sequence mutations only, or no IKZF1 alterations in whom favorable subtypes were enriched at 67.4%, 45%, and 59.3%, respectively (P<0.001). IK6 was observed in 16 patients, 11 of whom (68.8%) had the IKZF1plus profile.
Patients with IKZF1 alterations had poorer responses to induction therapy with EOI MRD ≥ 0.01% detected in 51.7% of patients with IKZF1plus, 22.2% of patients with IKZF1 deletions only, and 18.2% of patients with IKZF1 sequence mutations, compared to 12.5% of patients without IKZF1 alterations (P < 0.0001). EOI MRD ≥ 0.01% was observed in 56.3% of patients with IK6 and 32.6% of patients with non-IK6 deletions (P<0.0001).
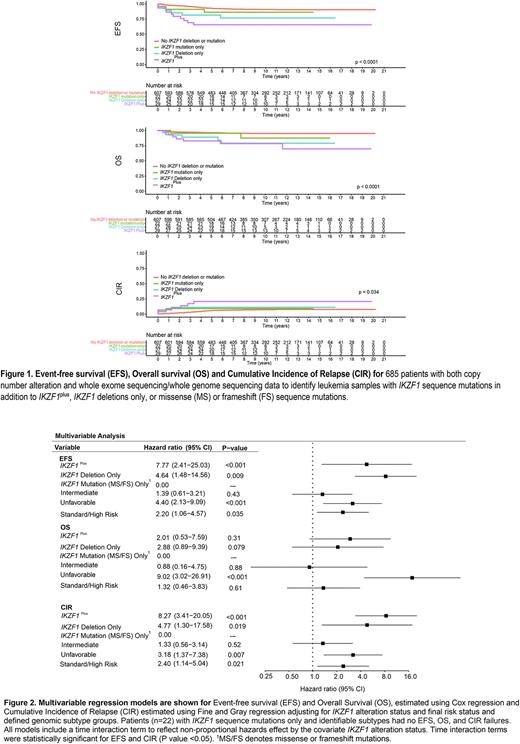
Patients with IKZF1plus had the poorest 5-year event free survival (EFS), overall survival (OS) and cumulative incidence of relapse (CIR) of 65.5 ± 8.8%, 82.8 ± 7.0%, and 20.7 ± 7.7%, respectively. IKZF1 deletions only were associated with EFS of 81.5 ± 7.5%, OS of 88.9 ± 6.1%, and CIR of 11.1 ± 6.2%. IKZF1 sequence mutations were associated with better outcomes than other IKZF1 alterations (EFS of 86.1 ± 7.4%, OS 95.5 ± 4.4% and CIR 9.1 ± 6.3). Patients without any IKZF1 alterations had the best clinical outcomes with EFS of 92.6 ± 1.1%, OS 96.3 ± 0.8%, and CIR 5.7 ± 1% (P<0.001 for comparison of all groups). Compared to patients with non-IK6 deletions, mutations only, or no deletions, the IK6 deletion conferred increased risk of relapse with EFS of 68.8 ± 12% and CIR 25 ± 11% (P<0.0001 for EFS and 0.03 for CIR).
Multivariable analyses, which included variables of genomic subtypes and final risk status, demonstrated an independent prognostic effect of both IKZF1plus and IKZF1 deletions only with the highest hazard ratio (HR) in EFS and CIR associated with IKZF1plus (EFS HR 7.77; 95% CI 2.41-25.03; P<0.001 and CIR HR 8.27; 95% CI 3.41-20.05); P<0.001). IKZF1 deletions only were associated with EFS HR 4.64; 95% CI 1.48 - 14.56; P<0.05 and CIR HR 4.77; 95% CI 1.30 - 17.58; P<0.05). IKZF1 sequence mutations were not associated with adverse outcomes. Unfavorable subtypes, including Ph and Ph-like-CRLF2 subtypes, had a significantly adverse prognostic effect.
Conclusions We show adverse independent prognostic effects of IKZF1plus and non-IKZF1plusIKZF1 deletions in patients treated in Total XV and XVI studies, adding to the importance of these genetic profiles in identifying patients at risk of relapse. Identification of genomic subtypes associated with IKZF1plus together with incorporation of sequence analysis to reveal the full spectrum of IKZF1 alterations further informs prognostication of B-ALL and reveals subgroups of patients who remain at risk of poor outcomes.
Disclosures
Inaba:Servier: Other: Grants and Personal Fees; Amgen: Other: Grants and Personal Fees; Incyte: Other: Grants; JAZZ: Other: Personal Fees; Chugai: Other: Personal Fees. Mullighan:Abbvie: Research Funding; Pfizer: Research Funding; Amgen: Honoraria; Illumina: Honoraria; Consulting: Honoraria; BEAM: Honoraria; FAZE: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal