Abstract
Master regulators, such as the hematopoietic transcription factor (TF) GATA1, play an essential role in orchestrating lineage commitment and differentiation. However, the precise mechanisms by which such TFs regulate transcription through interactions with specific cis-regulatory elements remain incompletely understood. Here, we describe a form of congenital hemolytic anemia caused by missense mutations in an intrinsically disordered region of GATA1, with a poorly understood role in transcriptional regulation. Through integrative functional approaches, we demonstrate that these mutations perturb GATA1 transcriptional activity by partially impairing nuclear localization and selectively altering precise chromatin occupancy by GATA1. These alterations in chromatin occupancy and concordant chromatin accessibility changes alter faithful gene expression, with failure to both effectively silence and activate select genes necessary for effective terminal red cell production. We demonstrate how disease-causing mutations can reveal regulatory mechanisms that enable the faithful genomic targeting of master TFs during cellular differentiation.
Key Points
Novel hemolytic anemia with elevated erythrocyte adenosine deaminase levels is associated with missense mutations (p.R307C/H) in GATA1.
Transcription of target genes is selectively altered because of disruption of faithful chromatin occupancy of GATA1 mutants.
Introduction
Master transcription factors (TFs) play fundamental roles in defining cell identity and enabling activation of specific cell-state maintenance or differentiation programs.1,2 These master regulators modulate gene expression by binding to cis-regulatory elements that function as enhancers, promoters, or silencers and often alter local and long-range chromatin architecture through such interactions.3,4 However, it is increasingly clear that most master regulators have numerous roles both within a specific cell type and across the range of cell states that occur during differentiation.4,5
GATA1 was among the first identified master regulators of hematopoiesis.6,7 In mice, GATA1 is necessary for production of red blood cells (RBCs; erythropoiesis), platelets (megakaryopoiesis), basophils, eosinophils, and mast cells.8 In addition, Gata1 has been shown to be sufficient to reprogram alternative hematopoietic lineages to an erythroid fate,9,10 demonstrating its potency to alter cell identity. Recent studies in humans have uncovered a range of blood disorders resulting from germline and somatic mutations in GATA1, which is encoded on the X chromosome. These mutations include those that are somatically acquired in the myeloproliferative disorder in infants with Down syndrome, leading to sole production of a short isoform of GATA1 lacking its N-terminal transactivation domain.6,11 Similar, although distinct, germline mutations that reduce the production of the full-length transactivation domain–containing version of GATA1 can result in Diamond-Blackfan anemia, with a paucity of erythroid precursors and progenitors in the bone marrow.12,13 A number of other distinct mutations in the N-terminal zinc finger cause altered differentiation and maturation of erythroid and megakaryocytic precursors.6,14,15 In all cases, the mutations result in impaired hematopoietic differentiation and perturb a major function of GATA1 in enabling hematopoiesis. Moreover, altered production of GATA1 as a result of impaired translation or splicing can compromise hematopoiesis.13,16-18 Notably, these mutations cause a diverse range of disorders that likely reflect impairment of specific aspects of hematopoietic differentiation or lineage commitment. Despite the considerable advances made through studies of such mutations and from other extensive functional studies,7 the mechanisms that enable precise genomic targeting of GATA1 during various stages of hematopoiesis remain incompletely understood.14
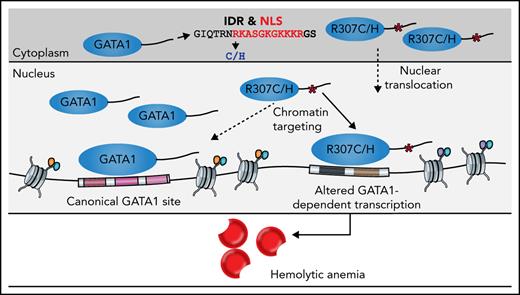
More than 40 years ago, a form of hemolytic anemia was first described and shown to involve altered nucleotide metabolism, with greatly increased levels of adenosine deaminase (ADA) produced within mature RBCs and an apparent autosomal dominant mode of inheritance.19 Several other cases have subsequently been characterized, including a family with possible X-linked inheritance by Miwa et al,20 another family with potential autosomal dominant inheritance by Pérignon et al,21 and a distinct family with apparent X-linked inheritance by our groups.22 However, the possible genetic and molecular bases of this type of hemolytic anemia have remained elusive.23,24 Here, we describe X-linked inheritance of this hemolytic anemia in families we and others have previously described20,22,25 that is attributable to mutations of arginine 307 in GATA1. Through functional studies, we demonstrate how these likely causal mutations disrupt a poorly characterized and intrinsically disordered C-terminal region of GATA1 that is essential for nuclear localization and precise chromatin occupancy by GATA1. We reveal a novel role for this C-terminal region of GATA1 to enable precise DNA binding, which when disrupted leads to a failure of effective terminal erythropoiesis and causes a hemolytic phenotype, thereby extending the range of erythroid disorders associated with GATA1 mutations and advancing our understanding of GATA1 function.
Methods
Patients, families, and cell biological procedures and methods
GATA1 R307C/H mutations were identified in 3 families with congenital hemolytic anemia via exome and targeted panel sequencing methodologies. Primary human bone marrow mononuclear cells were collected under institutional review board–approved protocols at Boston Children's Hospital, and written informed consent was received before inclusion in the study. Patient- and healthy control–derived hematopoietic cells were in vitro differentiated toward the erythroid lineage using established protocols. Complementary G1E and G1E-ER4 cells were used to model the effects of GATA1 mutations upon lentiviral complementation using complementary DNA expression vectors, followed by cell biological and biochemical characterization of cells. Detailed information about cell biological and biochemical approaches, including culture conditions, lentiviral vectors and transduction, flow cytometric analysis and sorting, cytoplasmic and nuclear fractionation, cycloheximide chase experiments, western blotting, coimmunoprecipitation and mass spectrometry, and quantitative reverse transcription polymerase chain reaction are provided in the data supplement.
Genomic and sequencing approaches
For comprehensive genomic characterization of primary erythroid human cells and cell lines expressing wild-type (WT) or mutant GATA1 protein, analyses of transcriptional, chromatin accessibility, and GATA1 chromatin occupancy profiles were undertaken using RNA sequencing (RNA-seq), assay for transposase-accessible chromatin sequencing (ATAC-seq), chromatin immunoprecipitation sequencing (ChIP-seq), CUT&RUN, and single-cell RNA-seq/ATAC-seq. Experimental details and bioinformatic analyses, including statistical details, are provided in the data supplement.
Results
Characterization of a distinct hemolytic anemia identifies C-terminal GATA1 mutations
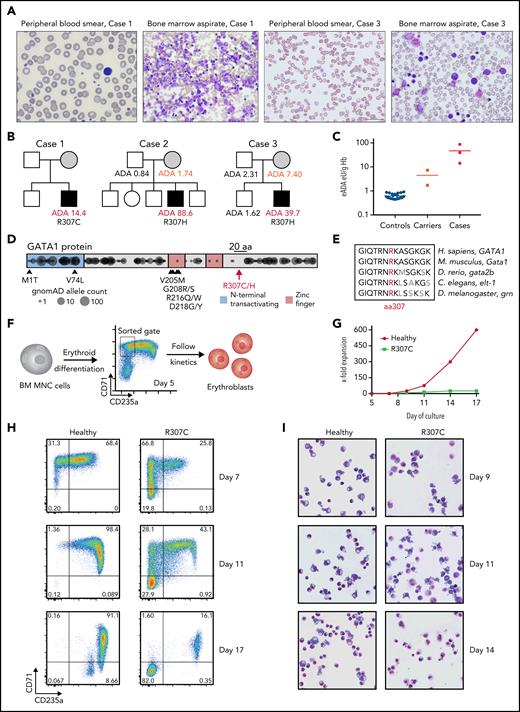
In the course of our studies of rare blood disorders,17,20,21,25-27 we identified 3 families where individuals were affected by hemolytic anemia characterized by shortened RBC lifespan, altered RBC morphology, increased reticulocyte production, and bone marrow erythroid hyperplasia with otherwise normal maturation of other blood cell lineages, consistent with the resultant compensation from RBC hemolysis22,25 (Figure 1A; supplemental Figure 1A; data supplement). Interestingly, all patients had high RBC erythrocyte adenosine deaminase (eADA) levels (Figure 1B-C), and the mothers in the cases seemed to have mildly elevated eADA, consistent with the previous suggestion of potential X-linked inheritance.20,22 A workup for typical causes of hemolysis in these cases was unrevealing, including a search for RBC membrane or enzyme disorders,28 and therefore, genomic sequencing was undertaken. In all patients, we identified previously unreported missense mutations at a highly conserved residue in the C-terminus of GATA1 (p.R307C/chrX:48652248 C>T and p.R307H/chrX:48652249 G>A in hg19), consistent with the posited X-linked inheritance (Figure 1B,D-E). Both mutations were predicted to be damaging (PolyPhen-2 score, 0.998; CADD PHRED score, >30)29,30 and were in a region belonging to the 97th highest percentile of constrained coding regions in the genome,31 and no mutations within 10 residues (297-317) have been observed in the gnomAD database (version 2.1.1).32
Patients with R307C/H GATA1 mutations display elevated ADA levels and impaired erythropoiesis. (A) Images of May-Grünwald-Giemsa–stained peripheral blood smears and bone marrow (BM) aspirates of indicated cases. (B) Pedigrees of 3 families with cases of congenital hemolytic anemia and elevated erythrocyte ADA (eADA) levels. Measured eADA levels are indicated. In all cases, GATA1 point mutations affecting Arg307 (R307) were identified. (C) Distribution of eADA levels in healthy controls (n = 21), suspected carriers (n = 2), and cases (n = 3). (D) Depiction of reported mutations in GATA1. Previously reported pathogenic variants are indicated with arrows and corresponding amino acid (aa) changes. R307 is in the middle of a 20-aa (black bar) region near the C-terminus. Circles represent projection of variants and their allele count as identified in ostensibly healthy individuals (n = 141 456; gnomAD). DNA binding zinc finger domains and the region around R307 are depleted for variants, indicating coding constraint. (E) aa conservation near R307 for indicated selected species. (F) Schematic of primary cell experiment. BM mononuclear cells (MNCs) from healthy donors and R307C mutant cells were in vitro differentiated into erythroblasts and sorted (gate as indicated), and differentiation and proliferation kinetics were monitored. (G) X-fold expansion of sorted healthy donor and R307C primary cells over the indicated time frame. (H) Flow cytometric plots of CD71 and CD235a surface marker–stained populations of healthy donor and R307C primary cells examined at indicated time points of culture. (I) Images of May-Grünwald-Giemsa–stained cytospin preparations of healthy donor and R307C primary cells at indicated time points of culture (×63 original magnification). Hb, hemoglobin.
Patients with R307C/H GATA1 mutations display elevated ADA levels and impaired erythropoiesis. (A) Images of May-Grünwald-Giemsa–stained peripheral blood smears and bone marrow (BM) aspirates of indicated cases. (B) Pedigrees of 3 families with cases of congenital hemolytic anemia and elevated erythrocyte ADA (eADA) levels. Measured eADA levels are indicated. In all cases, GATA1 point mutations affecting Arg307 (R307) were identified. (C) Distribution of eADA levels in healthy controls (n = 21), suspected carriers (n = 2), and cases (n = 3). (D) Depiction of reported mutations in GATA1. Previously reported pathogenic variants are indicated with arrows and corresponding amino acid (aa) changes. R307 is in the middle of a 20-aa (black bar) region near the C-terminus. Circles represent projection of variants and their allele count as identified in ostensibly healthy individuals (n = 141 456; gnomAD). DNA binding zinc finger domains and the region around R307 are depleted for variants, indicating coding constraint. (E) aa conservation near R307 for indicated selected species. (F) Schematic of primary cell experiment. BM mononuclear cells (MNCs) from healthy donors and R307C mutant cells were in vitro differentiated into erythroblasts and sorted (gate as indicated), and differentiation and proliferation kinetics were monitored. (G) X-fold expansion of sorted healthy donor and R307C primary cells over the indicated time frame. (H) Flow cytometric plots of CD71 and CD235a surface marker–stained populations of healthy donor and R307C primary cells examined at indicated time points of culture. (I) Images of May-Grünwald-Giemsa–stained cytospin preparations of healthy donor and R307C primary cells at indicated time points of culture (×63 original magnification). Hb, hemoglobin.
Impaired erythropoiesis and rescue experiments reveal altered activity of the GATA1 R307C/H mutants
To better define the cellular defect, we characterized erythroid cells derived from in vitro culture of bone marrow mononuclear cells from a patient harboring the GATA1 R307C mutation. Compared with healthy hematopoietic progenitor cells, the patient cells showed impaired differentiation to form CD71+CD235a+ erythroblasts, loss of surface marker expression during continued culture, reduced proliferation, and altered morphology (Figure 1F-I; supplemental Figure 1B-E), suggesting an impaired erythroid gene regulatory program of R307C mutant cells.
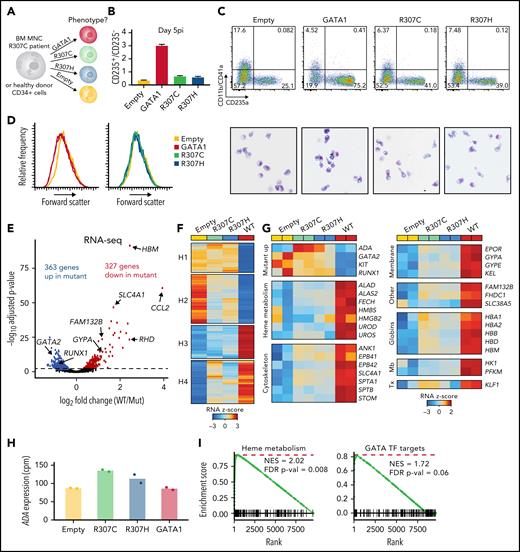
To experimentally assess the activity of mutant (R307C/H) compared with WT GATA1, complementary DNA constructs coupled to an internal ribosome entry site–green fluorescent protein (IRES-GFP) to confirm similar transduction efficiency were introduced via lentiviral transduction into patient and healthy donor primary cells (Figure 2A; supplemental Figure 2A). Exogenous expression of either R307C/H or GATA1 WT improved differentiation, as assessed by surface marker expression, morphology (more erythroid maturation observed), and forward scatter (decrease in cell size is indicative of more differentiated cells13; Figure 2B-D). Notably, GATA1 WT promoted differentiation more robustly, in both patient (Figure 2B-D) and healthy donor cells (supplemental Figure 2B-C). We conducted RNA-seq to assess transcriptional profiles of R307C patient cells exogenously expressing various GATA1 constructs, which further confirmed equivalent transduction efficacy between constructs based on IRES-GFP–derived reads (supplemental Figure 2D). GATA1 WT led to 549 up- and 546 downregulated genes compared with the empty vector control (adjusted P < .01) (supplemental Figure 2F). In contrast, the R307C/H mutants resulted in differential induction of a subset of these genes, 363 of which were up- and 327 downregulated compared with GATA1 WT (adjusted P < .01; Figure 2E; supplemental Table 1), with the gene expression changes between the R307C and R307H mutants relative to GATA1 WT being highly concordant (supplemental Figure 2E). K-means clustering further revealed distinct activity for different sets of genes, including genes that failed to be repressed (clusters H1 and H2) and genes that were not properly upregulated in GATA1 mutant transduced cells compared with GATA1 WT (clusters H3 and H4; Figure 2F). Clusters H1 and H2 contained genes related to myeloid and leukocyte activation (supplemental Table 3), respectively, as well as early hematopoietic progenitor genes, including GATA2, KIT, and RUNX1, which failed to be fully repressed in R307C/H-expressing cells (Figure 2G). In line with elevated eADA levels, we observed higher ADA messenger RNA expression in R307C/H transduced cells, which seemed to be specifically induced by the mutants, given the increase over GATA1 WT and the empty vector control (Figure 2G-H). In contrast, cluster H3 and H4 genes contained GATA1 target genes involved in key aspects of terminal erythroid maturation, including heme metabolism (H3) and 1 carbon metabolism (H4) alongside membrane proteins and hemoglobin genes (Figure 2G,I; supplemental Table 3). These findings suggest that the R307C/H mutants may be impaired in their ability to silence early hematopoietic and alternative fate genes, but they also show residual hypomorphic activity that is compatible with erythroid maturation, consistent with the clinical presentation of the patients.
Rescue with GATA1 WT improves erythroid differentiation and gene expression in primary R307C cells. (A) Schematic of primary cell rescue experiment. Indicated expression vectors were introduced to R307C patient bone marrow (BM) mononuclear (MNC) or healthy donor cells via lentiviral integration followed by in vitro culture. (B) Ratio of CD235+/CD235− cells as determined by flow cytometry of R307C cells transduced with indicated expression constructs after 5 days postinfection (pi). Error bars represent 1 standard error of the mean. (C) Flow cytometric plots (top row) of CD11b/CD41a and CD235a surface marker–stained populations and images of May-Grünwald-Giemsa–stained cytospin preparations (bottom row; ×63 original magnification) of R307C cells transduced with indicated expression constructs after 5 days pi. (D) Histogram plots of forward scatter of R307C cells transduced with indicated expression constructs assessed after 5 days pi. (E) Volcano plot showing differentially expressed genes comparing R307C cells transduced with mutant (R307C/H) with GATA1 WT–expressing vectors. Selected genes are indicated. Two replicates for GATA1 WT and all 4 mutant replicates were pooled for this analysis. (F) Heatmap showing results of k-means clustering of differentially expressed genes of in vitro cultured R307C patient cells transduced with indicated GATA1 constructs. Results of 2 replicates for each construct were pooled for this analysis. Color bar: z-scored expression. (G) z-score–transformed expression abundances based on RNA-seq for indicated GATA1 genotypes and selected genes with associated biological functions. Individual replicate results are depicted in the heatmap (2 per construct). (H) Bar graph showing ADA messenger RNA expression levels for indicated GATA1 genotypes. Individual replicates results are shown (dots; 2 replicates per construct). (I) Gene set enrichment analyses for indicated gene sets for differential expression analysis comparing R307C cells transduced with mutant and GATA1 WT. Two replicates for GATA1 WT and all 4 mutant replicates were pooled for this analysis. cpm, count per million; FDR, false discovery rate; Mb, metabolism; NES, normalized enrichment score; Tx, transcription.
Rescue with GATA1 WT improves erythroid differentiation and gene expression in primary R307C cells. (A) Schematic of primary cell rescue experiment. Indicated expression vectors were introduced to R307C patient bone marrow (BM) mononuclear (MNC) or healthy donor cells via lentiviral integration followed by in vitro culture. (B) Ratio of CD235+/CD235− cells as determined by flow cytometry of R307C cells transduced with indicated expression constructs after 5 days postinfection (pi). Error bars represent 1 standard error of the mean. (C) Flow cytometric plots (top row) of CD11b/CD41a and CD235a surface marker–stained populations and images of May-Grünwald-Giemsa–stained cytospin preparations (bottom row; ×63 original magnification) of R307C cells transduced with indicated expression constructs after 5 days pi. (D) Histogram plots of forward scatter of R307C cells transduced with indicated expression constructs assessed after 5 days pi. (E) Volcano plot showing differentially expressed genes comparing R307C cells transduced with mutant (R307C/H) with GATA1 WT–expressing vectors. Selected genes are indicated. Two replicates for GATA1 WT and all 4 mutant replicates were pooled for this analysis. (F) Heatmap showing results of k-means clustering of differentially expressed genes of in vitro cultured R307C patient cells transduced with indicated GATA1 constructs. Results of 2 replicates for each construct were pooled for this analysis. Color bar: z-scored expression. (G) z-score–transformed expression abundances based on RNA-seq for indicated GATA1 genotypes and selected genes with associated biological functions. Individual replicate results are depicted in the heatmap (2 per construct). (H) Bar graph showing ADA messenger RNA expression levels for indicated GATA1 genotypes. Individual replicates results are shown (dots; 2 replicates per construct). (I) Gene set enrichment analyses for indicated gene sets for differential expression analysis comparing R307C cells transduced with mutant and GATA1 WT. Two replicates for GATA1 WT and all 4 mutant replicates were pooled for this analysis. cpm, count per million; FDR, false discovery rate; Mb, metabolism; NES, normalized enrichment score; Tx, transcription.
Selectively altered transcriptional activity of GATA1 mutants in murine G1E cells
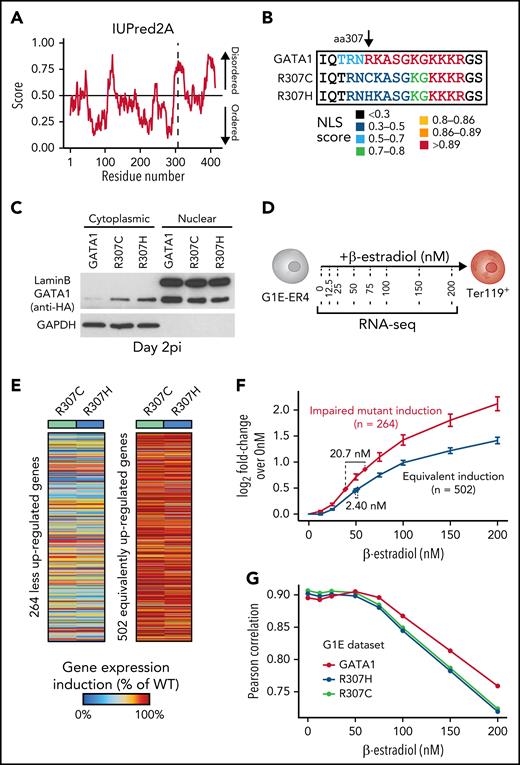
We next turned to Gata1−/− G1E cells, an established complementation system for the study of GATA1 function, which are derived from Gata1-knockout mouse pluripotent cells.33,34 Untagged and hemagglutinin-tagged GATA1 WT were lentivirally transduced with similar efficiency and led to the robust induction of Ter119, a marker that is upregulated during erythroid differentiation (Figure 3A-B; supplemental Figure 3A-B). In contrast, the R307C/H mutants led to reduced Ter119 induction. Importantly, GATA1 WT and mutant messenger RNA and protein levels were equivalent, and protein stability appeared unaltered (supplemental Figure 3C-E).35 Consistent with our primary cell data, RNA-seq analysis revealed altered activity of the R307C/H mutants. Of 1213 upregulated genes (GATA1 WT vs empty control), there was a select group of 138 up- and 329 genes that were downregulated (R307C/H vs GATA1 WT; adjusted P < .01; Figure 3C; supplemental Figure 3F-G; supplemental Table 1). Analogous to the human rescue data (Figure 2), k-means clustering revealed that the mutants displayed distinct activity for different sets of genes (Figure 3D; supplemental Figure 3H). Cluster K1 and K2 genes were readily repressed by GATA1 WT and mutants and contained alternate fate genes related to leukocyte (cluster K1) and platelet activation (cluster K2; supplemental Table 3). With respect to induced genes, we observed increased (cluster K3), reduced (cluster K4), or little induction (cluster K5) of genes when comparing R307C/H mutant with GATA1 WT activity (Figure 3D). The genes in all 3 clusters are normally upregulated during human erythropoiesis36 and include key regulators of terminal erythroid maturation (Figure 3E; supplemental Figure 3I). Pathway analysis of cluster K3 and K5 genes did not indicate an association with relevant biological processes but confirmed an enrichment of cluster K4 genes associated with heme biosynthesis and hemolytic anemia that are highly expressed during terminal erythropoiesis (Figure 3F; supplemental Table 3).37 Moreover, ATAC-seq38 revealed that the increase in chromatin accessibility at known GATA1 chromatin occupancy sites39 was delayed in mutant-expressing cells, with progressive increases in the observed differences in accessibility as cells underwent further maturation (Figure 3G). However, we observed that cluster- and gene-specific changes were less prominent (Figure 3H), suggesting that alterations in chromatin accessibility do not explain the pronounced differences at the transcriptional level in G1E cells (Figure 3D). To further assess the validity of the G1E complementation experiments, we conducted a cross-species correlation analysis, comparing the fold changes in gene expression by GATA1 WT relative to the mutants in the primary human R307C cells (Figure 2) and the murine G1E cells (Figure 3). Notably, the gene expression changes were highly concordant, suggesting cross-species conserved function of the C-terminal domain surrounding the R307C/H mutations (Figure 3I), in line with its evolutionary sequence preservation (Figure 1E). These results demonstrate that R307C/H display selectively altered transcriptional activity at canonical GATA1 target genes in human and mouse cells.
Differential transcriptional and epigenomic activity of mutant R307C/H and GATA1 WT. (A) Schematic of experiment. Indicated expression vectors were introduced to murine G1E cells via lentiviral integration followed by in vitro culture. (B) Frequency of Ter119+ cells at indicated day postinfection (pi) as assessed by flow cytometry. (C) Volcano plot of differentially expressed genes as determined by RNA-seq comparing G1E cells expressing mutant and GATA1 WT at day 3 pi. Selected genes are highlighted. (D) Heatmap showing results of k-means clustering of differentially expressed genes in G1E cells expressing indicated GATA1 constructs. Results of 2 replicates for each construct were pooled for this analysis. Color bar: z-scored expression. (E) Mean expression (log2 counts per million [cpm]) of genes per cluster from panel D across the process of human erythroid differentiation, including hematopoietic stem cells (HSCs), multipotent progenitor cells (MPPs), common myeloid progenitor (CMPs), megakaryocyte erythroid progenitor cells (MEPs), myeloid progenitor cells (MyPs), colony-forming unit erythroid cells (CFU-Es), proerythroblasts (ProE1 and ProE2), basophilic erythroblasts (BasoEs), polychromatic erythroblasts (PolyEs), orthochromatic erythroblasts (OrthoEs), and OrthoEs further enriched with reticulocytes (Orth/Ret). (F) Gene ontology (GO) terms for cluster K4 from panel D that identify key pathways associated with impaired gene induction by G1E cells expressing R307C/H compared with GATA1 WT. (G) Differences in chromatin accessibility as determined by ATAC-seq in G1E cells expressing indicated GATA1 constructs compared with GATA1 chromatin occupancy at respective sites in G1E-ER4 cells at indicated hours after differentiation induction. (H) Meta gene pileup analysis of chromatin accessibility patterns of genes in clusters K1 to K4 as determined in panel D. The position relative to the transcription start site (TSS) is shown. (I) Cross-species analysis of differential expression patterns of genes differentially regulated in the primary patient cell rescue data (Figure 2) and the G1E complementation experiments. The dot plot shows the log2 fold change (FC) for each gene comparing GATA1 WT relative to R307C/H mutant transduced cells for all genes that were differentially expressed in both species (adjusted P < .1; n = 226). Two replicates for GATA1 WT and all 4 mutant replicates were pooled for this analysis. BP, biological process; MF, molecular function.
Differential transcriptional and epigenomic activity of mutant R307C/H and GATA1 WT. (A) Schematic of experiment. Indicated expression vectors were introduced to murine G1E cells via lentiviral integration followed by in vitro culture. (B) Frequency of Ter119+ cells at indicated day postinfection (pi) as assessed by flow cytometry. (C) Volcano plot of differentially expressed genes as determined by RNA-seq comparing G1E cells expressing mutant and GATA1 WT at day 3 pi. Selected genes are highlighted. (D) Heatmap showing results of k-means clustering of differentially expressed genes in G1E cells expressing indicated GATA1 constructs. Results of 2 replicates for each construct were pooled for this analysis. Color bar: z-scored expression. (E) Mean expression (log2 counts per million [cpm]) of genes per cluster from panel D across the process of human erythroid differentiation, including hematopoietic stem cells (HSCs), multipotent progenitor cells (MPPs), common myeloid progenitor (CMPs), megakaryocyte erythroid progenitor cells (MEPs), myeloid progenitor cells (MyPs), colony-forming unit erythroid cells (CFU-Es), proerythroblasts (ProE1 and ProE2), basophilic erythroblasts (BasoEs), polychromatic erythroblasts (PolyEs), orthochromatic erythroblasts (OrthoEs), and OrthoEs further enriched with reticulocytes (Orth/Ret). (F) Gene ontology (GO) terms for cluster K4 from panel D that identify key pathways associated with impaired gene induction by G1E cells expressing R307C/H compared with GATA1 WT. (G) Differences in chromatin accessibility as determined by ATAC-seq in G1E cells expressing indicated GATA1 constructs compared with GATA1 chromatin occupancy at respective sites in G1E-ER4 cells at indicated hours after differentiation induction. (H) Meta gene pileup analysis of chromatin accessibility patterns of genes in clusters K1 to K4 as determined in panel D. The position relative to the transcription start site (TSS) is shown. (I) Cross-species analysis of differential expression patterns of genes differentially regulated in the primary patient cell rescue data (Figure 2) and the G1E complementation experiments. The dot plot shows the log2 fold change (FC) for each gene comparing GATA1 WT relative to R307C/H mutant transduced cells for all genes that were differentially expressed in both species (adjusted P < .1; n = 226). Two replicates for GATA1 WT and all 4 mutant replicates were pooled for this analysis. BP, biological process; MF, molecular function.
The GATA1 R307C/H mutants lie in an intrinsically disordered region
We sought to further analyze the region harboring the R307C/H mutations (Figure 1D-E), which has previously been suggested to be posttranslationally modified and implicated in protein interactions.40-42 The mutations are located within an intrinsically disordered region (IDR) of low sequence complexity, as has been described in a number of TFs (Figure 4A; supplemental Figure 4A).43,44 IDRs have been suggested to have essential functions in transcriptional regulation, including enabling multivalent interactions that facilitate formation of phase condensates and directing/modulating in vivo TF binding specificity.45-50 Because interactions may be critical for IDR function, we performed coimmunoprecipitation followed by mass spectrometry analyses (supplemental Figure 4B-D). This revealed conserved interactions with well-known binding partners, including components of the nucleosome remodeling deacetylase complex and ZFPM1 (supplemental Figure 4E-F).6,51,52 We observed only 18 differentially bound proteins of 939 detected proteins (adjusted P < .01; supplemental Figure 4G-H), one of which, GZF1, had a known role in transcriptional regulation53 and was depleted from R307C/H protein. However, repression of Gzf1 by CRISPR interference in G1E-ER4 cells, a G1E subclone that constitutively expresses an estradiol-activated form of GATA1 fused to the estrogen receptor ligand binding domain, enabling the ligand-dependent nuclear translocation and activation of GATA1-dependent transcription,39,54 did not affect Ter119 upregulation (supplemental Figure 4I-K; data supplement). These findings suggest that loss of GZF1 interactions were unlikely to have a major role in the observed transcriptional defects.
Localization of R307C/H in an IDR with a role in nuclear localization and GATA1 dosage-dependent target gene expression. (A) Computational prediction of IDRs across the full GATA1 protein using the indicated algorithm. The dotted line represents the R307 residue. Predictions from 5 additional algorithms also all indicate that the residue is part of a disordered domain (supplemental Figure 4A). (B) Nuclear localization signal annotation and score for amino acid (aa) sequences in R307C/H mutants and GATA1 WT. (C) Western blot of cytoplasmic and nuclear fractions of G1E cells expressing indicated GATA1 constructs at day 2 postinfection (pi). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and LaminB were used as cytoplasmic and nuclear markers to validate purity of fractionation. (D) Schematic of experiment. G1E-ER4 cells were incubated with increasing β-estradiol concentrations to titrate GATA1 nuclear translocation, transcriptional activity, and erythroid differentiation as measured by Ter119+ surface marker expression and RNA-seq. (E) Depiction of 2 gene sets with impaired (left; n = 264) and equivalent (right; n = 502) induction of gene expression in R307C/H mutant compared with GATA1 WT transduced G1E cells (from Figure 3). Color bar: gene expression induction (percentage of GATA1 WT). (F) Log2 fold change (FC) for each gene set as in panel E is shown for each β-estradiol–dependent RNA-seq sample from panel D. Error bars represent 1 standard error of the mean. Annotated points indicate the equivalent β-estradiol concentration representing the log2 FC for each gene set as observed in R307C/H and GATA1 WT transduced G1E cells as in panel E and Figure 3. (G) Correlation of gene expression profiles for R307C/H and GATA1 WT transduced G1E cells compared with variable β-estradiol level–treated G1E-ER4 cells. The mean Pearson correlation per population is shown. NLS, nuclear localization signal.
Localization of R307C/H in an IDR with a role in nuclear localization and GATA1 dosage-dependent target gene expression. (A) Computational prediction of IDRs across the full GATA1 protein using the indicated algorithm. The dotted line represents the R307 residue. Predictions from 5 additional algorithms also all indicate that the residue is part of a disordered domain (supplemental Figure 4A). (B) Nuclear localization signal annotation and score for amino acid (aa) sequences in R307C/H mutants and GATA1 WT. (C) Western blot of cytoplasmic and nuclear fractions of G1E cells expressing indicated GATA1 constructs at day 2 postinfection (pi). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and LaminB were used as cytoplasmic and nuclear markers to validate purity of fractionation. (D) Schematic of experiment. G1E-ER4 cells were incubated with increasing β-estradiol concentrations to titrate GATA1 nuclear translocation, transcriptional activity, and erythroid differentiation as measured by Ter119+ surface marker expression and RNA-seq. (E) Depiction of 2 gene sets with impaired (left; n = 264) and equivalent (right; n = 502) induction of gene expression in R307C/H mutant compared with GATA1 WT transduced G1E cells (from Figure 3). Color bar: gene expression induction (percentage of GATA1 WT). (F) Log2 fold change (FC) for each gene set as in panel E is shown for each β-estradiol–dependent RNA-seq sample from panel D. Error bars represent 1 standard error of the mean. Annotated points indicate the equivalent β-estradiol concentration representing the log2 FC for each gene set as observed in R307C/H and GATA1 WT transduced G1E cells as in panel E and Figure 3. (G) Correlation of gene expression profiles for R307C/H and GATA1 WT transduced G1E cells compared with variable β-estradiol level–treated G1E-ER4 cells. The mean Pearson correlation per population is shown. NLS, nuclear localization signal.
Reduced GATA1 nuclear localization alters transcription and mimics R307C/H mutants
Given the largely preserved protein interactions, and although we cannot exclude altered interactions at individual loci, we assessed whether specific sequence motifs in the IDR may be disrupted by the mutations. We noted that the GATA1 R307C/H mutations partially disrupted a predicted nuclear localization signal55 and observed that this mutation resulted in ∼40% reduction of GATA1 in the nucleus and increased GATA1 retention in the cytoplasm (Figure 4B-C; supplemental Figure 5A-B). To investigate the impact of reduced nuclear localization, we treated G1E-ER4 cells with increasing β-estradiol concentrations, thereby titrating nuclear GATA1 activity, as validated by changes in Ter119 expression (Figure 4D; supplemental Figure 5C-D). We then conducted RNA-seq to relate variation in nuclear GATA1 levels to R307C/H mutant transcriptional activity. For this comparison, we curated 2 gene sets of poorly and similarly induced genes in R307C/H compared with GATA1 WT transduced G1E cells (Figures 3 and 4E; supplemental Table 1; data supplement). Interestingly, the set of poorly induced genes was more highly transcribed at equivalent β-estradiol concentrations in G1E-ER4 cells, suggesting a higher degree of transcriptional responsiveness to GATA1 levels (Figure 4F; supplemental Figure 5E-G; supplemental Table 1). To estimate equivalent effect sizes, we projected the fold changes for each gene set as observed in GATA1 WT and R307C/H reconstituted G1E cells (Figure 3). This revealed the equivalent β-estradiol dose for the poorly induced gene set to be significantly lower for the R307C/H mutants (mean, 38.8 nM) compared with GATA1 WT (59.5 nM; Figure 4F). Considering all differentially expressed genes, R307C/H transduced G1E cells also showed greater correlation at lower β-estradiol concentrations (Figure 4G). Therefore, alterations of nuclear GATA1 levels may explain some of the observed changes in gene expression in R307C/H cells, although this is unlikely to fully explain the distinct transcriptional effects observed for different groups of GATA1 target genes (supplemental Figure 5G), some of which are more highly upregulated in R307C/H mutant reconstituted G1E cells (Figure 3D; cluster K3).
Single-cell genomic analyses reveal altered transcription and chromatin accessibility in primary patient cells
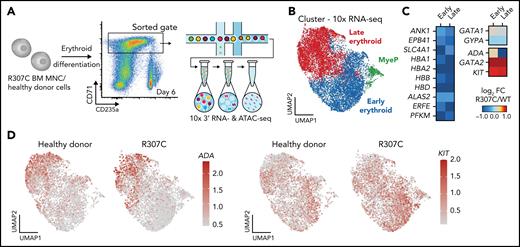
To gain further mechanistic insights at higher resolution, we conducted single-cell RNA-seq in patient and healthy donor cells. This analysis confirmed deregulated expression of key erythroid genes in stage-matched populations, with a notable failure to upregulate key genes involved in erythroid terminal maturation, along with a concomitant failure to silence a number of early progenitor-associated genes, including GATA2 and KIT (Figure 5A-D; supplemental Figure 6A-B). Complementary single-cell ATAC-seq analysis of these primary cells revealed altered chromatin accessibility that was consistent with the observed changes in gene expression (Figure 5C). Notably, we observed both decreases (supplemental Figure 6D-E) and increases in chromatin accessibility observed in R307C cells (supplemental Figure 6F). These observations suggest that R307C/H may lead to impaired or altered chromatin occupancy of GATA1 and subsequent impairment of chromatin structure and transcriptional regulation at key erythroid target genes.
Altered gene expression and chromatin accessibility in R307C GATA1 mutant primary hematopoietic cells. (A) Schematic of experiment. In vitro cultured bone marrow–derived R307C cells and healthy donor cells were sorted using the indicated gate at day 6 of culture for 10× single-cell RNA-seq and single-cell ATAC-seq processing. (B) Uniform manifold approximation and projection (UMAP) depicting myeloid progenitor (MyeP) and early and late erythroid compartments based on single-cell RNA-seq expression data. Healthy donor and R307C cells are jointly embedded, compared with supplemental Figure 6A. (C) Heatmap showing log2 fold change (FC; R307C/WT) of the expression of select genes as determined by single-cell RNA-seq analysis for indicated genes and cell cluster, compared with panel B. (D) UMAP as in panel B depicting ADA (left) and KIT (right) expression resolved by donor ID. BM, bone marrow; FC, fold change; MNC, mononuclear cell.
Altered gene expression and chromatin accessibility in R307C GATA1 mutant primary hematopoietic cells. (A) Schematic of experiment. In vitro cultured bone marrow–derived R307C cells and healthy donor cells were sorted using the indicated gate at day 6 of culture for 10× single-cell RNA-seq and single-cell ATAC-seq processing. (B) Uniform manifold approximation and projection (UMAP) depicting myeloid progenitor (MyeP) and early and late erythroid compartments based on single-cell RNA-seq expression data. Healthy donor and R307C cells are jointly embedded, compared with supplemental Figure 6A. (C) Heatmap showing log2 fold change (FC; R307C/WT) of the expression of select genes as determined by single-cell RNA-seq analysis for indicated genes and cell cluster, compared with panel B. (D) UMAP as in panel B depicting ADA (left) and KIT (right) expression resolved by donor ID. BM, bone marrow; FC, fold change; MNC, mononuclear cell.
Differential chromatin occupancy of GATA1 mutants
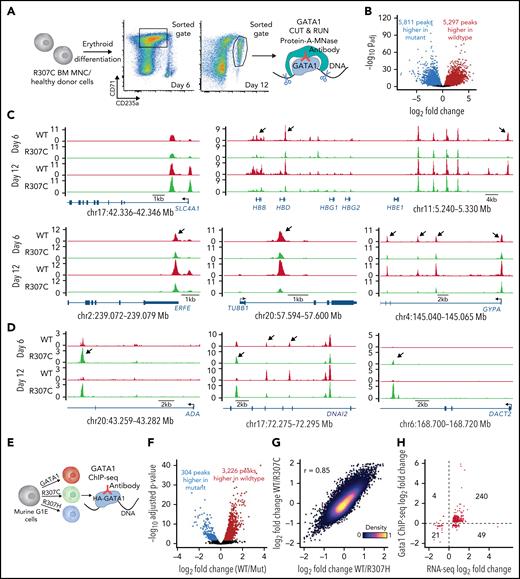
The R307 mutants lie within an IDR (Figure 4A; supplemental Figure 4A), and such regions have been shown to affect in vivo TF binding fidelity and the ability to target specific loci.49,50 We therefore sought to directly investigate whether the R307C substitution would alter GATA1 chromatin occupancy in stage-matched R307C patient and healthy control erythroblasts using the CUT&RUN approach (Figure 6A).56 Although ∼70% to 80% of GATA1 peaks were shared between patient and control samples, we observed notable chromatin occupancy patterns (Figure 6B-D; supplemental Figure 7A-B). For example, comparable binding near the SLC4A1 promoter was observed (Figure 6C, top left). In contrast, we noted significantly reduced binding at other key erythroid loci (Figure 6C, right and bottom), such as in the HBB locus. However, there was largely intact chromatin occupancy in the adjacent β-globin locus control region enhancer. Curiously, we also observed increased binding of the R307C mutant in specific regions (Figure 6D), including in the ADA locus, the expression of which was also elevated in these patients (Figure 1B-C). Overall, changes were largely correlated between the two assayed time points (Figure 6C-D). Moreover, these changes were concordant with the altered changes in chromatin accessibility we observed through single-cell ATAC-seq (supplemental Figure 6D-F). A 3-modal integrated analysis of the CUT&RUN, single-cell ATAC-seq, and single-cell RNA-seq data derived from the in vitro cultured primary R307C erythroblasts compared with healthy donor cells further revealed the high concordance of relative fold changes at the individual peak occupancy level and the chromatin accessibility and expression level of the closest genes at 498 sites at a false discovery rate of <0.1 for all 3 modalities (supplemental Figure 7C). Notably, we observed a markedly lower dynamic range in chromatin accessibility relative to GATA1 occupancy or gene expression, suggesting that altered transcription resulting from differential binding of the TF may occur on an already accessible landscape, more so than GATA1 mutants pioneering new accessible chromatin, consistent with our results in G1E cells (Figure 3).
Altered fidelity of GATA1 mutant chromatin occupancy in primary hematopoietic and G1E cells. (A) Schematic of experiment. In vitro cultured bone marrow (BM)–derived R307C cells and healthy donor cells were sorted using indicated gates at day 6 and day 12 of culture for GATA1 CUT&RUN. (B) Volcano plot showing differential GATA1 chromatin occupancy peaks in R307C and healthy donor cells (combined day 6 and day 12). (C-D) GATA1 chromatin occupancy at indicated loci in R307C and healthy donor cells at indicated time points, showing predominantly reduced (C) or increased (D) chromatin occupancy of the R307C mutant. Arrows highlight selected sites of differential chromatin occupancy. (E) Schematic of experiment. G1E cells transduced with indicated hemagglutinin (HA)-tagged GATA1 expression constructs were processed for ChIP-seq. (F) Volcano plot showing differential GATA1 chromatin occupancy peaks comparing G1E cells expressing R307C/H mutants and GATA1 WT. (G) Density plot showing correlation of chromatin occupancy fold changes between G1E cells expressing R307C or R307H mutant and GATA1 WT. (H) Correlation of chromatin occupancy and gene expression fold changes between G1E cells expressing R307C/H mutant and GATA1 WT. MNC, mononuclear cell.
Altered fidelity of GATA1 mutant chromatin occupancy in primary hematopoietic and G1E cells. (A) Schematic of experiment. In vitro cultured bone marrow (BM)–derived R307C cells and healthy donor cells were sorted using indicated gates at day 6 and day 12 of culture for GATA1 CUT&RUN. (B) Volcano plot showing differential GATA1 chromatin occupancy peaks in R307C and healthy donor cells (combined day 6 and day 12). (C-D) GATA1 chromatin occupancy at indicated loci in R307C and healthy donor cells at indicated time points, showing predominantly reduced (C) or increased (D) chromatin occupancy of the R307C mutant. Arrows highlight selected sites of differential chromatin occupancy. (E) Schematic of experiment. G1E cells transduced with indicated hemagglutinin (HA)-tagged GATA1 expression constructs were processed for ChIP-seq. (F) Volcano plot showing differential GATA1 chromatin occupancy peaks comparing G1E cells expressing R307C/H mutants and GATA1 WT. (G) Density plot showing correlation of chromatin occupancy fold changes between G1E cells expressing R307C or R307H mutant and GATA1 WT. (H) Correlation of chromatin occupancy and gene expression fold changes between G1E cells expressing R307C/H mutant and GATA1 WT. MNC, mononuclear cell.
To substantiate the observed altered binding of the GATA1 mutants, we used ChIP-seq using hemagglutinin-tagged GATA1 constructs in G1E cells (Figure 6E-H; supplemental Figure 8A). Again, we observed loci where chromatin occupancies of R307C/H compared with GATA1 WT appeared reduced, unaltered, or increased (supplemental Figure 8B-D). Furthermore, these differences in chromatin occupancy of the R307C/H mutants were largely concordant with the direction of changes in gene expression (Figure 6H; 83%; P < 2.2e−16; binomial test). Although we have attempted to identify particular motifs57 or other features that would distinguish sites that are differentially bound, including overlap with cell-cycle phase–dependent histone modifications58 and DNA shape,59 no clearly defined differences have been identified that would explain the observed DNA binding disparities (supplemental Figure 8E-G; supplemental Table 2). In addition, training of high-performance neural networks60-62 to predict CUT&RUN or ChIP-seq read profiles from the underlying DNA sequence and subsequent interrogation of these models also did not reveal clear distinctions between the R307C/H and GATA1 WT binding signatures. Our findings are therefore most consistent with the critical role of IDRs within TFs to facilitate precise chromatin targeting and modulate DNA binding by the TFs.47-50
Discussion
Here, we describe and functionally characterize the pathophysiology of a rare form of hemolytic anemia, with a phenotype distinct from those of other diseases caused by GATA1 mutations. This disorder primarily affects the production and function of mature RBCs, while ostensibly sparing earlier functions of GATA1 in hematopoiesis.6,17 Mild thrombocytopenia was also observed, resulting from either impaired megakaryopoiesis, as has been seen with other GATA1 mutants,14 or reticuloendothelial sequestration, as occurs in hemolytic anemias. Through studies of these newly characterized mutants, we show how a conserved C-terminal IDR in GATA1 plays an important and previously unappreciated role in enabling appropriate nuclear localization and faithful chromatin occupancy. Notably, the mutants seem to affect a select group of GATA1 chromatin occupancy sites, particularly those found in genes that are more dramatically upregulated at the later stages of erythropoiesis. This results in transcriptional deregulation of this specific group of genes, which includes key RBC membrane proteins, hemoglobin genes, heme biosynthetic enzymes, and other transcriptional regulators of terminal erythropoiesis. There is consequently impaired maturation of RBCs. Our findings are analogous to the selective transcriptional effects observed upon titration of glucocorticoid receptor nuclear localization.63 Although the improper silencing of early hematopoietic genes (eg, GATA2, RUNX1, and KIT) by the GATA1 mutants may contribute to the observed pathophysiology, the erythroid hyperplasia and reticulocyte response are consistent with a primary terminal erythroid defect, as opposed to impaired early erythroid commitment being the primary driver of disease, as is observed in other congenital anemias associated with GATA1 dysfunction, such as Diamond-Blackfan anemia.13,64 These clinical and molecular observations thereby illustrate the distinct and complex underlying pathophysiology of distinct forms of human disease associated with dysfunction of distinct aspects of the same GATA1 gene.
We attempted to identify particular DNA sequence motifs or other physical features that would distinguish sites that are differentially bound or target genes whose expression is affected by these mutants; however, no clearly defined differences were identified. Some of the observed transcriptional deficiencies at individual loci may be attributable to impaired GATA1 nuclear localization, although this seems insufficient to underlie all of the observed expression changes, including the increased or de novo chromatin occupancy by GATA1 observed at some loci. Considering the mitotic bookmarking capacity of GATA1 to occupy chromatin independent of cofactors such as FOG1 and TAL1, which are only recruited postmitotically,65 we suggest that the alterations observed may be due to a role for IDRs contributing to faithful TF binding specificity.49,50 Structural and biochemical studies will be required to fully elucidate the precise mechanisms of the disrupted protein-DNA binding interactions resulting from the mutated R307 residue and the contribution of the IDR that contains this residue to enable and modulate GATA1 chromatin occupancy.50,66 Independent of the role of IDRs to form transcriptional condensates via multivalent interactions, they may form a multitude of weak binding determinants across the IDR that may recognize broad DNA regions to direct TF binding specificity.49,50 In particular, short disordered segments flanking TF DNA binding domains have been shown to directly modulate specificity and affinity of DNA binding via interactions that localize to the DNA minor groove and that may not require specific sequences.50 However, independent of these observations and our proteomic analysis, we also cannot dismiss the additional contributions of altered cofactor-protein interactions at select loci, which will require more targeted studies using locus-specific interaction analyses.67,68
Overall, the molecular consequences of the R307C/H mutation are likely multifactorial, and it seems difficult to disentangle the individual contributions of altered nuclear localization, altered fidelity of chromatin occupancy, and locally perturbed protein interactions. However, the reported alterations provide a mechanistic understanding of how a unique phenotype can emerge as a result of these distinct GATA1 mutants, as normal silencing or activation of certain genes during terminal erythropoiesis fails to occur in the setting of these mutations. Future studies may provide deeper insight to help elucidate the exact molecular mechanisms contributing to ensure high-fidelity GATA1 chromatin interactions and the distinct requirements at different loci to explain the observed selectivity of affected sites.
In conclusion, our findings in the context of rare phenotype-altering alleles causing hemolytic anemia emphasize how appropriate gene expression during different stages of hematopoietic differentiation relies upon precise and effective chromatin occupancy by GATA1 via a key residue in the C-terminal IDR and point to the seldom-considered multifarious regulatory mechanisms that can exist for many master TFs, which when perturbed may lead to distinct human phenotypes.
Acknowledgments
The authors thank the patients and families for participating in the study, as well as the members of the Sankaran and Regev laboratories for valuable comments and the Whitehead Institute and Broad Institute Flow Cytometry facilities for assistance with cell sorting and flow cytometric analysis.
L.S.L. is supported by an Emmy Noether fellowship by the German Research Foundation (LU 2336/2-1) and a Hector Fellow Academy Research Career Development Award. C.A.L. received support from National Cancer Institute, National Institutes of Health (NIH), grant F31 CA232670 and a Stanford Science Fellowship and a Parker Institute of Cancer Immunotherapy Scholarship. E.L.B. received support from the Howard Hughes Medical Institute Medical Research Fellows program. This research was supported by National Institute of Diabetes and Digestive and Kidney Diseases, NIH, grant R01 DK103794 (V.G.S.), National Heart, Lung, and Blood Institute (NHLBI), NIH, grants R33 HL120791 (V.G.S.) and R01 HL146500 (V.G.S.), National Cancer Institute, grants U24 CA210986 (S.A.C.) and U01 CA214125 (S.A.C.); Japan Society for the Promotion of Science research grant KAKENHI JP16K10041 (H.K.), the Howard Hughes Medical Institute (A.R.), the Klarman Cell Observatory (A.R.), and the New York Stem Cell Foundation (NYSCF; V.G.S.). Funding for patient genomic sequencing was partially provided by the Broad Institute of Massachusetts Institute of Technology and Harvard Center for Mendelian Genomics and was funded by National Human Genome Research Institute, National Eye Institute, and National Heart, Lung, and Blood Institute grant UM1 HG008900 and by National Human Genome Research Institute grant R01 HG009141. V.G.S. is an NYSCF-Robertson Investigator.
Authorship
Contribution: L.S.L., C.A.L., T.U., T.L.M., H.K., and V.G.S. conceptualized the study; T.U., H.O., T.Y., H.I., S.N., S.O., A.H., B.G., R.C., T.L.M., H.K., and V.G.S. performed clinical assessments; L.S.L., C.A.L., E.L.B., A.M.T., N.L., S.A.M., W.L., C.F., M.E.O., C.M., C.M.V., M.M., V.S., A.R., and V.G.S. were responsible for methodology; C.A.L., E.L.B., L.S.L., A.M.T., J.M.V., N.L., J.C.U., and S.A.M. performed formal analyses, with input from C.M.V., S.A.C., M.J.A., A.R., and V.G.S; L.S.L., C.A.L., E.L.B., A.M.T., C.M., S.A.M., and C.M.V. performed investigations; S.A.C., M.J.A., A.K., A.R., and V.G.S. secured resources; L.S.L., C.A.L., E.L.B., and V.G.S. wrote, reviewed, and edited the original manuscript draft, with input from all authors; C.A.L., E.L.B., and L.S.L. were responsible for visualization; S.A.C., M.J.A., A.K., S.H.O., A.R., and V.G.S. supervised the study; and A.R. and V.G.S. were responsible for project oversight and funding acquisition.
Conflict-of-interest disclosure: J.C.U. has received compensation for consulting from Goldfinch Bio and is an employee of Patch Biosciences. C.A.L. is a consultant to Cartography Biosciences and SeQureDx. A.R. is a founder of and equity holder in Celsius Therapeutics, an equity holder in Immunitas Therapeutics, and until 31 August 2020 was a scientific advisory board member of Syros Pharmaceuticals, Neogene Therapeutics, Asimov, and Thermo Fisher Scientific. A.R. is an employee of Genentech (beginning 1 August 2020). V.G.S. serves as an advisor to and/or has equity in Branch Biosciences, Novartis, Forma, Cellarity, and Ensoma. The remaining authors declare no competing financial interests.
Leif S. Ludwig, Hannoversche St 28, 10115 Berlin, Germany; e-mail: leif.ludwig@bih-charite.de; and Vijay G. Sankaran, 1 Blackfan Circle, Karp Family Research Building Room 7211, Boston, MA 02115; e-mail: sankaran@broadinstitute.org.
REFERENCES
Author notes
Raw sequencing data related to this work is available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE133417.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
GATA1 is the master erythroid transcription factor, with inherited mutations linked to dyserythropoietic anemias such as Diamond-Blackfan anemia, and somatically acquired mutations linked to transient myeloproliferative disease in Down syndrome. Ludwig et al reveal that novel missense mutations in an intrinsically disordered region (IDR) in the carboxyl-terminal domain of GATA1 cause a rare form of X-linked inherited hemolytic anemia. In addition to explaining the molecular basis for this disease, the data shed light on the precise function of the IDR in target gene activation.
L.S.L. and C.A.L. contributed equally to this work.

![Differential transcriptional and epigenomic activity of mutant R307C/H and GATA1 WT. (A) Schematic of experiment. Indicated expression vectors were introduced to murine G1E cells via lentiviral integration followed by in vitro culture. (B) Frequency of Ter119+ cells at indicated day postinfection (pi) as assessed by flow cytometry. (C) Volcano plot of differentially expressed genes as determined by RNA-seq comparing G1E cells expressing mutant and GATA1 WT at day 3 pi. Selected genes are highlighted. (D) Heatmap showing results of k-means clustering of differentially expressed genes in G1E cells expressing indicated GATA1 constructs. Results of 2 replicates for each construct were pooled for this analysis. Color bar: z-scored expression. (E) Mean expression (log2 counts per million [cpm]) of genes per cluster from panel D across the process of human erythroid differentiation, including hematopoietic stem cells (HSCs), multipotent progenitor cells (MPPs), common myeloid progenitor (CMPs), megakaryocyte erythroid progenitor cells (MEPs), myeloid progenitor cells (MyPs), colony-forming unit erythroid cells (CFU-Es), proerythroblasts (ProE1 and ProE2), basophilic erythroblasts (BasoEs), polychromatic erythroblasts (PolyEs), orthochromatic erythroblasts (OrthoEs), and OrthoEs further enriched with reticulocytes (Orth/Ret). (F) Gene ontology (GO) terms for cluster K4 from panel D that identify key pathways associated with impaired gene induction by G1E cells expressing R307C/H compared with GATA1 WT. (G) Differences in chromatin accessibility as determined by ATAC-seq in G1E cells expressing indicated GATA1 constructs compared with GATA1 chromatin occupancy at respective sites in G1E-ER4 cells at indicated hours after differentiation induction. (H) Meta gene pileup analysis of chromatin accessibility patterns of genes in clusters K1 to K4 as determined in panel D. The position relative to the transcription start site (TSS) is shown. (I) Cross-species analysis of differential expression patterns of genes differentially regulated in the primary patient cell rescue data (Figure 2) and the G1E complementation experiments. The dot plot shows the log2 fold change (FC) for each gene comparing GATA1 WT relative to R307C/H mutant transduced cells for all genes that were differentially expressed in both species (adjusted P < .1; n = 226). Two replicates for GATA1 WT and all 4 mutant replicates were pooled for this analysis. BP, biological process; MF, molecular function.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/139/16/10.1182_blood.2021013753/6/m_bloodbld2021013753f3.jpeg?Expires=1769881400&Signature=QjBMxeyGAld84yT7v5YryICCCaqZDKmmxx4vSK8ZrN3-4UeDuVaBsM7jw~UPZ-fDHHJ2m1tFs~YJ-84K~TfP8tp4PJp8A1lkq8NI8gT8YiVkTna46IUGDVeuW65cJahJn6hCWkiAjoMk8lPG5cJ18tljilu2fIUcbSn7fqEiezwzGwoMcYCmU7701-IPmJy4h0ljlmPMX5FxPeElAgAEBy7edFhbsRSoElh-~VmPHeZT9nBAPt4BZbek4JBsmVMhirj~ykQvzW6OCsipXOdW8T-Cp-6S8SuVdI17m8aNOLmjg6ZB7ix-yol6oXa4ajK9IYVbqW4zjJComYmjuUgzHQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal