Abstract
Introduction
Despite recent improvement in outcomes for de novo disease, pediatric T-cell acute lymphoblastic leukemia (T-ALL) remains challenging to treat at relapse. Investigation into genomic markers of treatment response and therapy resistance offers an opportunity to further enhance outcomes for these patients. We previously identified a T-ALL blast-associated gene signature at diagnosis (Dx) and characterized the immune microenvironment in Dx T-ALL marrow samples using single cell transcriptome analysis (Bhasin et al. Blood 2020(ASH)). This approach allowed us to generate a granular expression map of both the T-ALL landscape and the Dx bone marrow (BM) immune microenvironment. Here we expand this work by evaluating samples collected from the same patients Dx and End of Induction (EOI) BM samples from pediatric T-ALL patients. The use of paired samples provides insight into treatment-induced changes in the microenvironment. Further, the inclusion of both minimal residual disease (MRD) positive and MRD negative samples allowed us to compare differences between these groups.
Methods
Using the 10X genomics platform, we profiled the single cell transcriptome of ~18,000 BM and immune microenvironment cells from viably frozen samples collected from T-ALL patients at Dx or EOI. Five paired Dx and EOI samples and one EOI sample from a patient with relapsed T-ALL were evaluated, for a total of 11 samples. Three paired samples were MRD positive at EOI and two were MRD negative; the relapsed sample was MRD negative. Cell clustering was performed using the Seurat package and differential expression analysis was performed using R/Bioconductor packages (Hao et al. Cell 2021). Cell communication analysis was conducted using the CellChat R tool (v 1.0.0) to infer cell-cell communication within the EOI MRD positive and MRD negative subsets and compare their communication networks (Jin et al. Nature Comm 2021).
Results
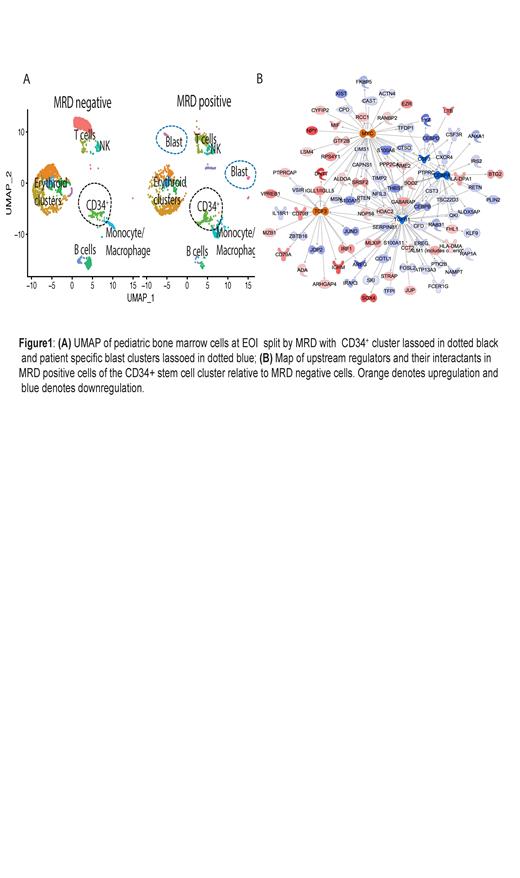
Using our previously described blast-associated gene signature (Bhasin et al. ASH 2020) we were able to identify residual blast populations at EOI in MRD-positive samples. Comparative analysis of gene profiles at Dx and EOI showed significant changes in the microenvironment cell populations with highest increase in erythroid cell populations after induction therapy. The gene expression profiles were significantly different for immune cells at Dx and EOI and the relapsed sample had greater similarity to the Dx samples indicating a persistent immunosuppressive environment. Clustering analysis of the EOI samples (3 MRD positive and 2 MRD negative) demonstrated the presence of patient specific blast cells in MRD positive samples that retained patient-specific transcriptomeheterogeneity at EOI (Fig.1A). Analysis of communication networks between different cell types based on receptor and ligand expression levels between different cell types identified a CD34 + cluster of stem cells that had different interactions with other immune populations in the MRD positive and negative subsets. Differential expression analysis between the MRD positive and MRD negative cells in this CD34 + stem cell cluster identified higher expression of myeloid associated genes such as CEBPB, CEBPD, AZU1 in the MRD negative group relative to the MRD positive cells, which showed higher expression of B-cell related genes such as IGHM, VPREB1, CD79A/ B along with upregulation of P13K signaling in B-lymphocytes, B-cell receptor signaling and autophagy pathways. Analysis of upstream regulators based on the differential gene signature between the MRD positive and MRD negative group demonstrated upregulation of MYC and TCF3 activity and inhibition of TGFB1, CSF3 and CEBPA in MRD positive compared to MRD negative samples (Fig.1B).
Conclusions: Leukemic blasts exhibit patient-specific gene expression signatures that are present at EOI in MRD positive samples. Exploration of the impact of minimal residual disease at EOI revealed differential gene expression patterns in stem cells from MRD positive samples, characterized by activation of B cell related signaling pathways and regulators such as MYC and TCF3. In contrast, a more myeloid-like expression signature was observed in stem cells from MRD negative samples. These findings open the avenues for exploration of therapeutic targets of T-ALL progression.
DeRyckere: Meryx: Other: Equity ownership. Graham: Meryx: Membership on an entity's Board of Directors or advisory committees, Other: Equity ownership.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal