In this issue of Blood, Sheng et al show that the N6-methyladenosine (m6A) messenger RNA (mRNA) reader protein YTH domain containing 1 (YTHDC1) promotes leukemic cell proliferation through stabilizing transcriptions of DNA replication initiation complex subunit MCM4.1
The central dogma of biology is that DNA is transcribed to RNA, which is then translated into protein. Each step of the process is highly regulated by discrete modifications to the target molecule and associated factors. These modifications impart changes in the target molecule so profound to its function that related mutations account for most oncogenic drivers. The field of cell biology has been broadly focused on two-thirds (DNA and protein) of the central dogma trifecta. We still need to understand how modifications to RNA contribute to disease and development.
RNA modifications affect the stability, localization, and translation efficiency of the molecule. The m6A modification cooccurs with transcription and occurs within a DRACH (A/G/U; G/A; A; C; A/C/U) consensus sequence.2 Broadly speaking, the METTL family of proteins are responsible for writing the mark,3 YTH domain-containing proteins such as YTHDC1 are responsible for reading the mark,4 and erasers such as FTO and ALKBH5 are responsible for removing the mark.5,6 In the past few years, several studies have highlighted the importance of these marking pathways in leukemogenesis. For example, 1 study showed that METTL3 (m6A writer) cooperates with CEBPZ to bind promoters of key acute myeloid leukemia (AML) survival genes and promote their translation,7 and other studies showed that m6A erasers FTO and ALKBH5 are necessary for AML development through regulation of RARA, MYC, and CEBPA.8,9
YTHDC1 is localized to cell cycle-associated YT bodies, commonly known as nuclear speckles, and is primarily responsible for facilitating the cytoplasmic export of m6A-marked mRNA. Although YTHDC1 has been shown to preferentially bind noncoding RNAs such as XIST,9 there are many mRNAs that are exported from the nucleus without YTHDC1 involvement. This suggests there are additional levels of regulation that remain to be uncovered, and studies like the one by Sheng et al present evidence for a novel role for YTHDC1 in promoting the stability of mRNA transcripts.
Sheng et al report the observation that YTHDC1 is broadly upregulated in AML samples compared with healthy hematopoietic tissues, and selective depletion of YTHDC1 through RNA interference in leukemic cell lines leads to growth arrest and loss of colony formation. They then confirm the intrinsic requirement of YTHDC1 to AML cell growth and survival with an exhaustive examination of many retrovirally expressed oncogene models including MLL-AF9, AML1-ETO9a, HOXA9, and PML-RARα. In each case, elevated YTHDC1 expression was required for growth and colony formation in vitro and disease development in vivo. Haploinsufficiency significantly impaired leukemic stem cell function in vitro and in vivo. This finding contrasted with the effects of YTHDC1 reduction in healthy hematopoiesis, in which there was no discernable phenotype in YTHDC1 haploinsufficient animals.
The most salient discovery in this work is the finding that YTHDC1 does not necessarily drive the nuclear export of key transcripts, but instead predominantly stabilizes the transcripts of genes related to cell cycle. This was surprising, as previous accounts of YTHCD1 function in other cell biology models primarily attribute YTHDC1 interactions to nuclear export.10 Using RNA sequencing to identify changes in transcript levels following YTHDC1 knockdown in the MOLM13 AML cell line, Sheng et al identified several components of the DNA replication initiation complex with reduced transcripts following YTHDC1 knockdown including members of the minichromosome maintenance (MCM) family MCM2/4/5, the anaphase promoting complex, and the chromatin assembly factor 1a. However, as these transcript levels could be changing because of cell cycle, Sheng et al used the RNA-binding protein immunoprecipitation assay to show their direct interaction of YTHDC1 was responsible for their stability. Knockdown of YTHDC1 in AML cell lines led to DNA damage and cell-cycle arrest, which the authors determined was due to defective replication initiation. In support of their findings, they found reexpression of MCM4 in YTHDC1 knockdown cells was sufficient to reverse the phenotype. The cell growth and colony formation defects were seen most profoundly in several primary patient AML samples when compared with the relatively little effects on healthy CD34+ peripheral blood stem cells from healthy donors.
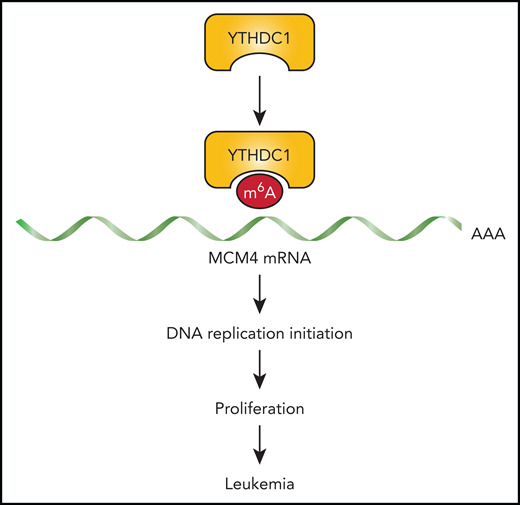
In summary, Sheng et al show us that YTHDC1 is a key leukemogenesis gene. They propose YTHDC1 reads the m6A mark on target transcripts (with the focus on MCM4 in this study) promoting its stability. In the case of AML, the transcript targets of YTHDC1 are broadly associated with the cell cycle, which results in supporting proliferation of the malignant clone. Acute leukemia-initiating cells capitalize on this activity because of their comparatively higher expression of YTHDC1 (see figure).
m6A reader protein YTHDC1 promotes leukemogenesis by stabilizing m6A-marked MCM4 mRNA. YTHDC1 is highly expressed in malignant hematopoietic cells, which contributes to preservation of MCM4 transcripts. This leads to stabilization of the replication initiation complex through adequate levels of MCM4, which in turn supports the rapid proliferation of malignant hematopoietic cells.
m6A reader protein YTHDC1 promotes leukemogenesis by stabilizing m6A-marked MCM4 mRNA. YTHDC1 is highly expressed in malignant hematopoietic cells, which contributes to preservation of MCM4 transcripts. This leads to stabilization of the replication initiation complex through adequate levels of MCM4, which in turn supports the rapid proliferation of malignant hematopoietic cells.
This study highlights the importance of mRNA reader proteins in the pathobiology of AML. And like all good studies, this one leaves us with many more questions. Some questions include exploring the basic biology into how YTHDC1 expression is regulated. Is there a leukemia-specific program that is promoting the upregulation of YTHDC1 in the malignant context that is different from healthy hematopoiesis? Downstream of YTHDC1, how is the protein promoting the stability of MCM4 in AML? Is it through preventing degradation by the nuclear PAXT complex, or are there other mechanisms? There are molecules identified that are directed toward inhibiting RNA methylation, but most inhibit either the writing or the erasing of the m6A mark. Could there be a way to target mRNA readers like YTHDC1 in a way that would selectively affect leukemic cells and spare healthy hematopoietic cells?
YTHDC1 is a conductor that allows AML to expand and make a mess on the metaphorical train of the bone marrow. Studies like this from Sheng et al have demonstrated that we may be able invalidate its ticket by targeting RNA methylation.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal