Key Points
Functional EpoR is expressed in subsets of tissue macrophages and hematopoietic cells.
EpoR-tdTomato-Cre mouse line provides a powerful tool for identification of EpoR expression and gene manipulation in EpoR-expressing cells.
Abstract
The erythropoietin receptor (EpoR) has traditionally been thought of as an erythroid-specific gene. Notably, accumulating evidence suggests that EpoR is expressed well beyond erythroid cells. However, the expression of EpoR in non-erythroid cells has been controversial. In this study, we generated EpoR-tdTomato-Cre mice and used them to examine the expression of EpoR in tissue macrophages and hematopoietic cells. We show that in marked contrast to the previously available EpoR-eGFPcre mice, in which a very weak eGFP signal was detected in erythroid cells, tdTomato was readily detectable in both fetal liver (FL) and bone marrow (BM) erythroid cells at all developmental stages and exhibited dynamic changes during erythropoiesis. Consistent with our recent finding that erythroblastic island (EBI) macrophages are characterized by the expression of EpoR, tdTomato was readily detected in both FL and BM EBI macrophages. Moreover, tdTomato was also detected in subsets of hematopoietic stem cells, progenitors, megakaryocytes, and B cells in BM as well as in spleen red pulp macrophages and liver Kupffer cells. The expression of EpoR was further shown by the EpoR-tdTomato-Cre–mediated excision of the floxed STOP sequence. Importantly, EPO injection selectively promoted proliferation of the EpoR-expressing cells and induced erythroid lineage bias during hematopoiesis. Our findings imply broad roles for EPO/EpoR in hematopoiesis that warrant further investigation. The EpoR-tdTomato-Cre mouse line provides a powerful tool to facilitate future studies on EpoR expression and regulation in various non-hematopoietic cells and to conditionally manipulate gene expression in EpoR-expressing cells for functional studies.
Introduction
Erythropoietin (EPO) and its cognate receptor, the erythropoietin receptor (EpoR), are indispensable for the proliferation, survival, and maturation of erythroid progenitors and early-stage erythroblasts.1-3 Mice lacking either EPO or EpoR die of severe anemia at embryonic day 13.3 The dependence of erythropoiesis on EPO/EpoR led to the initial notion that EpoR expression is restricted to erythroid lineage. However, accumulating evidence suggests broad expression of EpoR in non-erythroid cells.4-6 In the hematopoietic system, it has been reported that EPO stimulated bone morphogenetic protein expression by hematopoietic stem cells (HSCs),7 guided multipotent hematopoietic progenitor cells toward an erythroid fate,8 promoted differentiation of megakaryocytes,9 stimulated immunoglobulin production and proliferation of B cells,10,11 and promoted dying cell clearance by macrophages.12 Other well-documented non-erythropoietic roles of EPO include angiogenesis,13,14 neuroprotection,15-18 brain development,19-22 and metabolism/energy homeostasis.5 These functional studies strongly suggest EpoR expression in the related tissues and cell types. However, in many cases, the expression of EpoR has either been assessed only at the messenger RNA level23 or not examined at all, largely due to lack of monoclonal antibody for detecting surface expression of mouse EpoR.
Genetically engineered mouse models with knock-in of a fluorescent protein under the promoter of the gene of interest provide powerful tools for monitoring gene expression. Previously, Heinrich et al24 generated an EpoR-eGFPcre knockin mouse model by knocking in the eGFPcre at the EpoR locus. Specifically, the coding sequence of the eGFPcre was inserted in exon 1, replacing the translational start codon ATG of the EpoR by the translational start site of the eGFPcre complementary DNA, leading to replacement of the endogenous EpoR by the eGFPcre. Because the homozygous eGFPcre allele results in a lethal EpoR knock-out at the embryonic stage, the viable EpoR-eGFPcre mice are heterogeneous for EpoR and eGFPcre. Using this mouse line, the authors showed that weak enhanced green fluorescent protein (eGFP) fluorescence was detected in erythroid progenitors but not in other hematopoietic cells. Moreover, the EpoR-eGFPcre mice may be haploinsufficient because it has been reported that EpoR+/− mice are haploinsufficient.25
Here we found that the EpoR-eGFPcre mice were indeed haploinsufficient, raising the concern about use of this mouse line for functional studies. To address this issue, we generated an EpoR-tdTomato-Cre mouse line in which tdTomato and Cre were knocked in at the end of the EpoR coding sequence, and a P2A sequence was inserted between EpoR and tdTomato, as well as between tdTomato and Cre, to allow independent expression of EpoR, tdTomato, and Cre under the control of the EpoR promoter.26,27 We show that the EpoR-tdTomato-Cre mice had normal hematologic parameters. We further show that EpoR-tdTomato was expressed in subsets of tissue macrophages and hematopoietic cells. Importantly, EPO injection selectively promoted proliferation of the EpoR-expressing cells and induced erythroid lineage bias during hematopoiesis. Our findings imply previously unanticipated broad roles for EPO/EpoR in hematopoiesis. The EpoR-tdTomato-Cre mouse model should facilitate future studies of EPO/EpoR biology beyond hematopoiesis.
Materials and methods
Antibodies and mice
Antibodies used in this study are listed in supplemental Table 1 (available on the Blood Web site). EpoR-eGFPcre knockin mice were described previously.24 Gt(ROSA)26Sortm1(EYFP)Cos mice28 and Gt(ROSA)26Sortm14(CAG-tdTomato)Hze mice29 were purchased from The Jackson Laboratory. EpoR-tdTomato-Cre mice were generated by using CRISPR/Cas9 technology at Biocytogen. Animal protocols were reviewed and approved by the Institutional Animal Care and Use Committee of New York Blood Center and Zhengzhou University.
Blood parameter measurement and colony assay
Blood was analyzed by using an ADVIA 120 hematology analyzer (Siemens Healthineers). Burst-forming unit–erythroid (BFU-E) and colony-forming unit–erythroid (CFU-E) assays were performed in MethoCult GF M3434 or MethoCult M3334 (Stemcell Technologies) medium, respectively. Colonies were counted as previously described.30
Preparation of single-cell suspension, depletion of lineage+ cells
For bone marrow (BM), cells were flushed with phosphate-buffered saline (PBS)/2% fetal bovine serum/2 mM EDTA. For fetal liver (FL) or spleen, FL or spleen was placed into a cell strainer and smashed through the cell strainer into a petri dish. For liver, the shredded tissues were first digested with digestion buffer. For lineage+ cell depletion, cells were suspended at the concentration of 107/100 μL and incubated with biotin-conjugated anti-lineage antibodies, followed by incubation with anti-biotin beads, as described in detail in the supplemental Materials and methods.
Enrichment of macrophages, B cells, EPO treatment, and western blot
BM EpoR-tdTomato+ macrophages or B cells were sorted on a BD FACSAria Fusion cell sorter. Spleen and liver macrophages were enriched by using F4/80 beads. The cells were starved for 6 hours on ice, then stimulated with EPO for 10 minutes at 37°C. Details are described in the supplemental Materials and methods.
Staining and flow cytometric analysis of lineage– cells and erythroblasts
The lineage– cells were suspended at the concentration of 0.1 × 106/35 μL and stained with multiple antibodies. The erythroid cells in BM and FL were stained as previously described,31-33 with some modifications. The details of antibody usage and instrument voltage settings are described in the supplemental Materials and methods.
Flow cytometric analyses of macrophages, hematopoietic stem and progenitor cells, megakaryocytes, and lymphocytes
BM and FL macrophages were stained as described in our previous publication,34 with some modifications. Spleen and liver macrophages, BM hematopoietic stem, and progenitor cells, megakaryocytes, and B cells were stained by using the combination of known surface markers for these cell populations.35-39 Details are described in the supplemental Materials and methods.
Fluorescence-activated cell sorting
To sort F4/80+EpoR-tdTomato+ macrophages, CD19+EpoR-tdTomato+ B cells or BFU-E, CFU-E and erythroblasts, BM or FL single cells were stained as described in the supplemental Materials and methods. The cells were sorted on a BD FACSAria Fusion cell sorter.
Amnis imaging flow cytometry of native EBIs
Native mouse EBIs were prepared as previously described.34,40 The enriched EBIs were stained with fluorescein isothiocyanate–conjugated anti-mouse Ter119, AF647-conjugated anti-mouse F4/80, and Hoechst33342 at 37°C for 1 hour. The cells were analyzed by using an Amnis ImageStream Mark II instrument at ×60 magnification. IDEAS software was used to analyze imaging flow data.
Statistics
FlowJo software was used to analyze flow cytometry data; ImageJ was used to analyze band signal intensities; and GraphPad Prism software was used for statistical analysis. All data are reported as mean ± standard error of the mean. Differences among 2 groups were calculated by using an unpaired Student t test.
Results
EpoR-eGFPcre mice were haploinsufficient
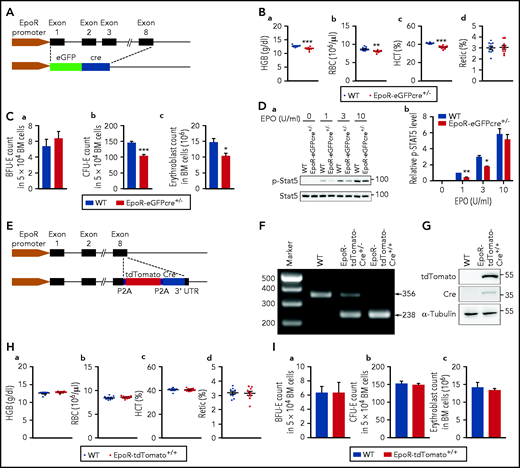
The EpoR-eGFPcre transgenic mouse line was generated 16 years ago24 and has been widely used for conditional gene deletion thought to be of erythroid lineage.41-50 However, due to the nature by which the EpoR-eGFPcre mouse line was generated (Figure 1A), it is likely that the EpoR-eGFPcre mice are haploinsufficient. To test this possibility, we measured blood parameters. Figure 1B shows that EpoR-eGFPcre mice exhibited mild but significant decreases in hemoglobin, red blood cell (RBC) count, and hematocrit. We also quantified erythroid colonies and erythroblasts in the BM. Although no difference was noted in the numbers of BFU-E colonies (Figure 1C-a), CFU-E colonies (Figure 1C-b) and erythroblasts (Figure 1C-c) were significantly decreased in the BM of the EpoR-eGFPcre mice compared with that of the littermate control. Furthermore, phosphorylation levels of Stat5 in EpoR-eGFPcre BM cells were significantly lower than that of the littermate control (Figure 1D), particularly at low EPO concentrations. These findings show that the EpoR-eGFPcre mice are haploinsufficient.
EpoR-eGFPcre mice were haploinsufficient. (A) Schematic model of EpoR-eGFPcre mouse model. (B) RBC parameters as indicated. N = 10. (C) (a) The number of BFU-E colonies in 5 × 104 BM cells. (b) The number of CFU-E colonies in 5 × 104 BM cells. (c) The number of erythroblasts in 2 tibia and 2 femur. N = 3. (D) Representative western blot (a) and quantitative results (b) of pSTAT5 levels. STAT5 level was used as loading control. N = 3. (E) Schematic model for generation of EpoR-tdTomato-Cre mice. (F) Polymerase chain reaction analysis showing the genotypes of EpoR-tdTomato-Cre mice. (G) western blot analysis showing the expression of tdTomato and Cre in BM cells of the EpoR-tdTomato-Cre mice. (H) Peripheral blood red cell parameters exhibiting no differences between EpoR-tdTomato-Cre mice and littermate control mice. N = 10. (I) BM erythroid colonies and erythroblasts showing no differences between EpoR-tdTomato-Cre mice and littermate control mice. N = 3. *P < .05; **P < .01; ***P < .001. HCT, hematocrit; HGB, hemoglobin; WT, wild type.
EpoR-eGFPcre mice were haploinsufficient. (A) Schematic model of EpoR-eGFPcre mouse model. (B) RBC parameters as indicated. N = 10. (C) (a) The number of BFU-E colonies in 5 × 104 BM cells. (b) The number of CFU-E colonies in 5 × 104 BM cells. (c) The number of erythroblasts in 2 tibia and 2 femur. N = 3. (D) Representative western blot (a) and quantitative results (b) of pSTAT5 levels. STAT5 level was used as loading control. N = 3. (E) Schematic model for generation of EpoR-tdTomato-Cre mice. (F) Polymerase chain reaction analysis showing the genotypes of EpoR-tdTomato-Cre mice. (G) western blot analysis showing the expression of tdTomato and Cre in BM cells of the EpoR-tdTomato-Cre mice. (H) Peripheral blood red cell parameters exhibiting no differences between EpoR-tdTomato-Cre mice and littermate control mice. N = 10. (I) BM erythroid colonies and erythroblasts showing no differences between EpoR-tdTomato-Cre mice and littermate control mice. N = 3. *P < .05; **P < .01; ***P < .001. HCT, hematocrit; HGB, hemoglobin; WT, wild type.
Generation and phenotypic characterization of EpoR-tdTomato-Cre mice
The finding that the EpoR-eGFPcre mice are haploinsufficient raises the concern for the use of this mouse line as a tool for functional studies. To address this issue, we generated an EpoR-tdTomato-Cre mouse line. As shown in the schematic model (Figure 1E), tdTomato and Cre were knocked-in at the end of the EpoR coding sequence, and a P2A sequence was inserted between EpoR and tdTomato, as well as between tdTomato and Cre, to allow independent expression of EpoR, tdTomato, and Cre under the control of the EpoR promoter. Genotype was confirmed by polymerase chain reaction (Figure 1F). Western blots show that tdTomato and Cre recombinase were expressed in BM cells from the EpoR-tdTomato-Cre mice but not from wild-type mice (Figure 1G). EpoR-tdTomato-Cre mice were born at the Mendelian ratio, viable, and did not exhibit any gross defects. Analyses of peripheral blood red cell parameters (Figure 1H), BM erythroid colonies, and erythroblasts (Figure 1I) revealed no differences between EpoR-tdTomato-Cre mice and the littermate control mice. These findings show that the EpoR is fully functional in the EpoR-tdTomato-Cre mice.
tdTomato but not eGFP was readily detected in FL and BM erythroid progenitors
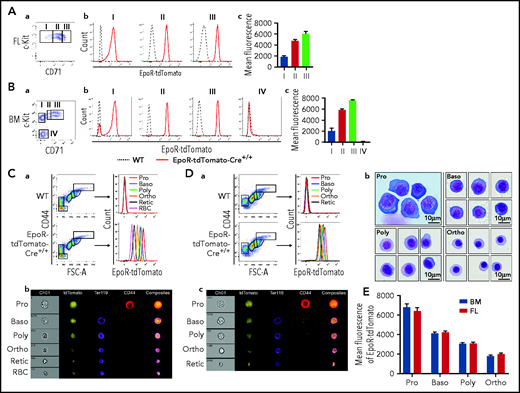
Erythropoiesis contains 8 morphologically and functionally distinct developmental stages: BFU-E, CFU-E, proerythroblast (Pro), basophilic erythroblast (Baso), polychromatic erythroblast (Poly), orthochromatic erythroblast (Ortho), reticulocyte (Retic), and RBC. We first examined the expression of EpoR in mouse FL BFU-E and CFU-E using the EpoR-tdTomato-Cre mice. According to the method developed by Flygare et al,31 FL BFU-E and CFU-E cells were defined as 10% Lin–c-Kit+CD71Low and 20% Lin–c-Kit+CD71High, respectively. Figure 2A shows that tdTomato was readily detected in the FL BFU-E (population I), CFU-E (population III), and the cell population in between (population II, which should be a mixture of BFU-E and CFU-E cells).
Expression of EpoR-tdTomato in mouse FL and BM erythroid cells. FL (A) and BM (B) progenitors. (a) Plot of CD71 vs c-Kit of 7AAD–Linages–CD16–CD32–CD41–CD34–Sca1– cells revealing 3 populations in FL (I: BFU-E; II: BFU-E/CFU-E mixture; and III: CFU-E) and 4 populations in BM (I: c-Kit+CD71–; II: c-Kit+CD71Low; III: c-Kit+CD71High; and IV: c-Kit–CD71–). (b) Histogram of EpoR-tdTomato showing EpoR-tdTomato expression levels in indicated cell populations. (c) Quantification of mean fluorescence intensity of EpoR-tdTomato. (C) BM erythroblasts. (a) Left panel: Representative plot of CD44 vs FSC-A of Ter119+ cells revealing different staged erythroid cells; right panel: histogram of EpoR-Tdtomato of the indicated cell population revealing progressive decrease of EpoR-tdTomato from Pro to RBC. (b) Representative images of IFC analysis showing expression of EpoR-tdTomato as well as changes in cell size, Ter119, and CD44 from Pro to RBC. (D) FL erythroblasts. (a) Left panel: Representative plot of CD44 vs FSC-A of Ter119+ cells revealing different staged erythroid cells. Right panel: histogram of EpoR-Tdtomato of the indicated cell populations. (b) Composite representative cytospin images of the sorted erythroblasts. (c) Representative images of IFC analysis. (E) Quantification of mean fluorescence intensity of EpoR-tdTomato. N = 3. FSC-A, forward scatter area; WT, wild type; IFC, imaging flow cytometry.
Expression of EpoR-tdTomato in mouse FL and BM erythroid cells. FL (A) and BM (B) progenitors. (a) Plot of CD71 vs c-Kit of 7AAD–Linages–CD16–CD32–CD41–CD34–Sca1– cells revealing 3 populations in FL (I: BFU-E; II: BFU-E/CFU-E mixture; and III: CFU-E) and 4 populations in BM (I: c-Kit+CD71–; II: c-Kit+CD71Low; III: c-Kit+CD71High; and IV: c-Kit–CD71–). (b) Histogram of EpoR-tdTomato showing EpoR-tdTomato expression levels in indicated cell populations. (c) Quantification of mean fluorescence intensity of EpoR-tdTomato. (C) BM erythroblasts. (a) Left panel: Representative plot of CD44 vs FSC-A of Ter119+ cells revealing different staged erythroid cells; right panel: histogram of EpoR-Tdtomato of the indicated cell population revealing progressive decrease of EpoR-tdTomato from Pro to RBC. (b) Representative images of IFC analysis showing expression of EpoR-tdTomato as well as changes in cell size, Ter119, and CD44 from Pro to RBC. (D) FL erythroblasts. (a) Left panel: Representative plot of CD44 vs FSC-A of Ter119+ cells revealing different staged erythroid cells. Right panel: histogram of EpoR-Tdtomato of the indicated cell populations. (b) Composite representative cytospin images of the sorted erythroblasts. (c) Representative images of IFC analysis. (E) Quantification of mean fluorescence intensity of EpoR-tdTomato. N = 3. FSC-A, forward scatter area; WT, wild type; IFC, imaging flow cytometry.
Next, we sought to assess the expression of EpoR in BM BFU-E and CFU-E. For this, we first examined whether the method for isolating FL BFU-E and CFU-E can also isolate BM BFU-E and CFU-E cells. The gating procedure is presented in supplemental Figure 1A, which shows that, different from FL, there was a c-Kit+CD71– population within the c-Kit+ fraction (population I). Within the c-Kit+CD71+ population, 2 clusters can be separated, c-Kit+CD71Low (population II) and c-Kit+CD71High (population III). Cytospin images of the sorted populations are presented in supplemental Figure 1B, which displays clear morphologic differences of these 3 populations. Supplemental Figure 1C shows that the BM c-Kit+CD71–, c-Kit+CD71Low, and c-Kit+CD71High cells predominantly gave rise to BFU-E colonies (with >70% purity), mixture of BFU-E/CFU-E colonies, and CFU-E colonies (with >90% purity), respectively. It is interesting to note that although both population I and II gave rise to BFU colonies, the sizes of colonies given by population I were generally bigger than those by population II, suggesting they are early and late BFU-E, respectively. Figure 2B shows that tdTomato was readily detected in all subpopulations of the c-Kit+ cells, although it was not detected in the c-Kit– population (population IV).
We next examined the expression of EpoR using the EpoR-eGFPcre mice. Supplemental Figure 2 shows that, in marked contrast to the clear detection of tdTomato in both FL and BM erythroid progenitor cells, eGFP was barely detected in these cells. This finding indicates that the weak fluorescence of the eGFP limits the use of the EpoR-eGFPcre mice for detection of EpoR expression.
Progressive changes of EpoR expression during terminal erythroid differentiation revealed by EpoR-tdTomato expression but not by EpoR-eGFP expression
During terminal erythroid differentiation, Pro sequentially differentiate to Baso, Poly, and Ortho. Ortho extrude the nucleus to become Retic, which mature into red cells in the blood stream. Our laboratory has documented that these 6 cell populations in BM can be clearly separated.33,Figure 2C-a shows that tdTomato was most abundantly expressed in Pro and decreased progressively, reflecting the progressive decrease of EpoR expression during terminal erythroid differentiation. Images from ImageStream analyses also show that tdTomato fluorescence was progressively decreased and that tdTomato was diffusely distributed in the cell (Figure 2C-b). Notably, tdTomato was also detected in reticulocytes and RBCs. Given the fact that reticulocytes/RBCs are reportedly devoid of EpoR51 and that tdTomato has a long half-life,52 we reasoned that a long half-life of tdTomato could account for the low level of tdTomato expression after EpoR is no longer expressed. To test this theory, we monitored the changes of tdTomato in peripheral blood RBCs with time. As expected, the fluorescence of tdTomato remained at the same level during the 20-day period (supplemental Figure 3).
We used the same method to examine the expression of EpoR-tdTomato in day 14.5 FL erythroid cells. Figure 2D-a shows that day 14.5 FL Ter119+ erythroid cells were clearly separated into 5 clusters. Cytospin images of the sorted erythroblast populations revealed that morphologically they resemble Pro, Baso, Poly, and Ortho, respectively (Figure 2D-b), showing that the method for isolating BM erythroblasts at the distinct developmental stage can be used to isolate FL erythroblasts as well. ImageStream analyses revealed progressive decease of tdTomato fluorescence from Pro to Retic (Figure 2D-c). Quantification of tdTomato fluorescence intensity revealed that similar levels of tdTomato were detected in BM and FL erythroblasts (Figure 2E).
We next used the EpoR-eGFPcre mice to examine the expression of EpoR in the terminally differentiated erythroid cells in both BM and FL. In contrast to the clear detection of tdTomato in erythroblasts, the fluorescence of eGFP in different staged erythroid cells was barely detectable and overlapped, failing to resolve progressive changes in EpoR expression (supplemental Figure 4A). Consistent with the flow cytometric analyses, ImageStream analyses also showed that the eGFP fluorescence in erythroblasts was faint (supplemental Figure 4B).
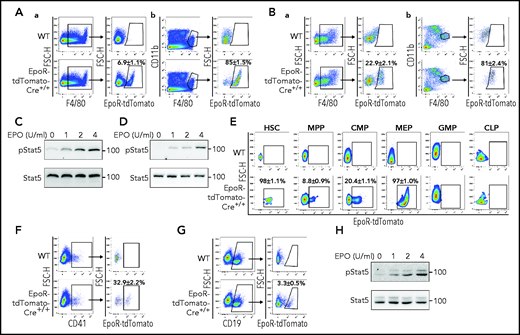
EpoR-tdTomato was expressed in subsets of BM and FL macrophages
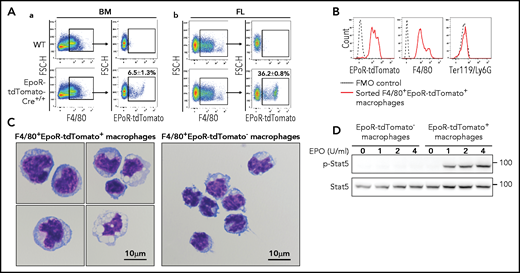
Using the EpoR-eGFPcre mice, we recently documented that EpoR-eGFPcre is expressed in subsets of BM and FL macrophages and that EBIs are predominantly formed by the F4/80+EpoR-eGFPcre+ macrophages.34 Here we further confirm the expression of EpoR in subsets of BM and FL macrophages using the EpoR-tdTomato-Cre mice. The gating procedure for the EpoR-tdTomato+ macrophages is essentially the same procedure used for gating the EpoR-eGFP+ macrophages in our recent publication,34 except that the SSCHigh population was gated out for further analysis (supplemental Figure 5). Figure 3A shows that, similar to F4/80+EpoR-eGFP+ macrophages, the F4/80+EpoR-tdTomato+ macrophages account for ∼6.5% of BM and ∼36% of day 14.5 FL F4/80+ cells, respectively. Flow cytometric analyses of the sorted BM F4/80+EpoR-tdTomato+ macrophages show that they express F4/80+EpoR-tdTomato+ but not erythroid marker Ter119 (Figure 3B), indicating that the observed EpoR-tdTomato expression in macrophages is not due to contamination by erythroid cells. Cytospin images of the sorted BM macrophages show that the F4/80+EpoR-tdTomato+ macrophages are morphologically distinct from the F4/80+EpoR-tdTomato– macrophages and that no erythroid cells are seen inside the macrophages (Figure 3C), indicating that detection of EpoR-tdTomato in macrophages is not due to phagocytosized erythroid cells. More cytospin images of the sorted BM macrophages are provided in supplemental Figure 6. Importantly, EPO trigged Stat5 phosphorylation in the sorted F4/80+EpoR-tdTomato+ macrophages but not in the F4/80+EpoR-tdTomato– macrophages (Figure 3D), providing direct evidence that F4/80+EpoR-tdTomato+ macrophages express EpoR on their surface.
Expression of EpoR-tdTomato in BM and FL macrophages. (A) Flow cytometric analyses of EpoR-tdTomato in BM (left panels) and day 14.5 FL (right panels) macrophages showing expression of EpoR-tdTomato in subsets of BM and FL F4/80+ macrophages. (B) Flow cytometric analyses of the sorted BM F4/80+EpoR-tdTomato+ macrophages showing expression of F4/80 and EpoR-tdTomato but not Ter119 or Ly6G. (C) Composite representative cytospin images of the sorted BM F4/80+EpoR-tdTomato+ and F4/80+EpoR-tdTomato– macrophages. Scale bar, 10 μm. (D) western blot analysis of pSTAT5 of the sorted macrophages stimulated with EPO showing dose-dependent phosphorylation of pSTAT5 in F4/80+EpoR-tdTomato+ but not F4/80+EpoR-tdTomato–macrophages. N = 3.
Expression of EpoR-tdTomato in BM and FL macrophages. (A) Flow cytometric analyses of EpoR-tdTomato in BM (left panels) and day 14.5 FL (right panels) macrophages showing expression of EpoR-tdTomato in subsets of BM and FL F4/80+ macrophages. (B) Flow cytometric analyses of the sorted BM F4/80+EpoR-tdTomato+ macrophages showing expression of F4/80 and EpoR-tdTomato but not Ter119 or Ly6G. (C) Composite representative cytospin images of the sorted BM F4/80+EpoR-tdTomato+ and F4/80+EpoR-tdTomato– macrophages. Scale bar, 10 μm. (D) western blot analysis of pSTAT5 of the sorted macrophages stimulated with EPO showing dose-dependent phosphorylation of pSTAT5 in F4/80+EpoR-tdTomato+ but not F4/80+EpoR-tdTomato–macrophages. N = 3.
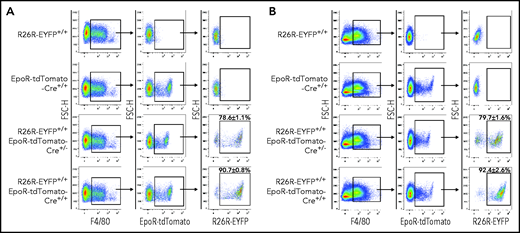
EpoR-tdTomato-Cre–mediated recombination in erythroid cells
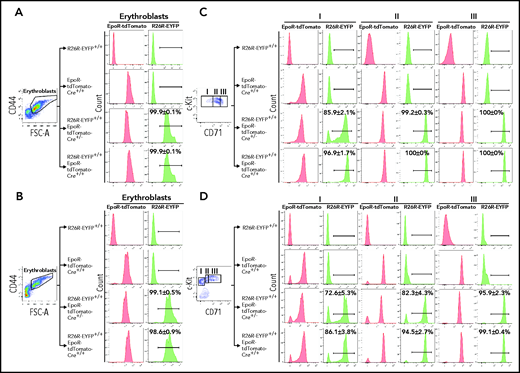
The EpoR-tdTomato-Cre mouse line was generated to serve 2 purposes, monitoring the expression of EpoR using the strong fluorescence of the tdTomato and conditional gene manipulation in EpoR-expressing cells in conjunction with the Cre-loxP recombination system. To examine the Cre activity of the EpoR-tdTomato-Cre mice, we crossed the EpoR-tdTomato-Cre+/+ mice with Gt(ROSA)26Sortm1(EYFP) Cos mice, also known as R26R-EYFP mice, which have a loxP-flanked STOP sequence followed by the EYFP inserted into the Gt(ROSA)26Sor locus.28 When bred to mouse expressing Cre recombinase, the STOP sequence is cleaved, and EYFP expression is observed in the Cre-expressing cells of the double-mutant offspring. As shown in Figure 4A-B, EpoR-tdTomato-Cre led to cleavage in almost 100% of both FL and BM erythroblasts, as shown by expression of R26R-EYFP in these cells. In contrast, the cleavage in erythroid progenitors seems to be dependent on the levels of Cre. While EpoR-tdTomato-Cre+/− led to cleavage in ∼86% and ∼73% of FL and BM population I cells, respectively, EpoR-tdTomato-Cre+/+ led to cleavage in ∼97% and ∼86% of the same population (Figure 4C-D). Moreover, although both EpoR-tdTomato-Cre+/− and EpoR-tdTomato-Cre+/+ led to the expression of R26R-EYFP in almost 100% of FL population II and III cells, EpoR-tdTomato-Cre+/− and EpoR-tdTomato-Cre+/+ led to the expression of R26R-EYFP in ∼82% and ∼94% of BM population II cells and ∼96% and ∼99% of BM population III.
EpoR-tdTomato-Cre–mediated cleavage in erythroid cells. FL (A) and BM (B) erythroblasts. Left panels: representative plots of FSC-A vs CD44 of Ter119+ cells. Erythroblasts were gated. Right panels: expression of EpoR-tdTomato or R26R-EYFP in erythroblasts of different mouse strains as indicated. FL (C) and BM (D) progenitors. Left panels: representative plots of c-Kit vs CD71 of 7AAD–Linages–CD16–CD32–CD41–CD34–Sca1– cells revealing 3 populations of the c-Kit+ cells. Right panels: expression of EpoR-tdTomato or R26R-EYFP in each erythroid progenitor population of different mouse strains as indicated. N = 3. FSC-A, forward scatter area.
EpoR-tdTomato-Cre–mediated cleavage in erythroid cells. FL (A) and BM (B) erythroblasts. Left panels: representative plots of FSC-A vs CD44 of Ter119+ cells. Erythroblasts were gated. Right panels: expression of EpoR-tdTomato or R26R-EYFP in erythroblasts of different mouse strains as indicated. FL (C) and BM (D) progenitors. Left panels: representative plots of c-Kit vs CD71 of 7AAD–Linages–CD16–CD32–CD41–CD34–Sca1– cells revealing 3 populations of the c-Kit+ cells. Right panels: expression of EpoR-tdTomato or R26R-EYFP in each erythroid progenitor population of different mouse strains as indicated. N = 3. FSC-A, forward scatter area.
We next examined cleavage by EpoR-eGFPcre in which Cre was fused with eGFP. For this analysis, we crossed the EpoR-eGFPcre mouse with the Cre reporter strain Gt(ROSA)26Sortm14(CAG-tdTomato)Hze mice.29 Although EpoR-eGFPcre+/− led to cleavage in almost 100% of erythroblasts, the cleavage in erythroid progenitors was less efficient (supplemental Figure 7A-B). Notably, R26R-tdTomato was only detected in ∼25% of FL population I cells (supplemental Figure 7C) and in ∼31% of BM population I cells, respectively (supplemental Figure 7D). This shows that the cleavage mediated by EpoR-eGFPcre+/− in population I cells is much lower than that by EpoR-tdTomato-Cre+/−, indicating that the Cre activity was affected by fusion with eGFP.
EpoR-tdTomato-Cre–mediated recombination in BM and FL EpoR+ macrophages
We next examined EpoR-tdTomato-Cre–mediated excision in macrophages. Figure 5 shows that EpoR-tdTomato-Cre+/− and EpoR-tdTomato-Cre+/+ led to cleavage in ∼80% and ∼90% of both FL and BM EpoR-tdTomato+ macrophages, respectively, as shown by the expression of R26R-EYFP in these cell populations.
EpoR-tdTomato-Cre mediated recombination in macrophages. FL (A) and BM (B). Left panels: representative plots of F4/80 vs forward scatter height (FSC-H) of Ly6G–Ter119– cells revealing F4/80+ macrophages; middle panels: representative plots of EpoR-tdTomato vs FSC-H of F4/80+ cells revealing EpoR-tdTomato+ macrophage population; right panel: representative plots of R26R-EYFP vs FSC-H of the EpoR-tdTomato+ macrophages revealing EpoR-Cre–mediated recombination efficiency in EpoR-tdTomato+ macrophages. N = 3.
EpoR-tdTomato-Cre mediated recombination in macrophages. FL (A) and BM (B). Left panels: representative plots of F4/80 vs forward scatter height (FSC-H) of Ly6G–Ter119– cells revealing F4/80+ macrophages; middle panels: representative plots of EpoR-tdTomato vs FSC-H of F4/80+ cells revealing EpoR-tdTomato+ macrophage population; right panel: representative plots of R26R-EYFP vs FSC-H of the EpoR-tdTomato+ macrophages revealing EpoR-Cre–mediated recombination efficiency in EpoR-tdTomato+ macrophages. N = 3.
tdTomato was readily detected in the surrounding erythroid cells of native EBIs from the EpoR-tdTomato-Cre mice
We recently documented that both mouse and human EBIs are predominantly formed by the EpoR+ macrophages.34 However, in analyzing the native EBIs of the EpoR-eGFPcre mice, it was difficult to detect eGFP fluorescence in the surrounding erythroid cells of most EBIs. In the current study, we found that this might be due to the weak fluorescence signal of the eGFP in erythroid cells. Having shown much stronger tdTomato fluorescence in erythroid cells, we expected that tdTomato would be readily detected in the surrounding erythroid cells of the native EBIs from the EpoR-tdTomato-Cre mice. As expected, tdTomato was indeed readily seen in both the central F4/80+ macrophages and the surrounding Ter119+ erythroblasts (supplemental Figure 8A). Quantitative analyses found that >90% of both FL and BM native EBIs were formed by the F4/80+EpoR-tdTomato-Cre+ macrophages (supplemental Figure 8B). Together, these results further confirm our recent finding that EBI macrophages are characterized by the expression of EpoR.
Functional EpoR is expressed in spleen red pulp macrophages and liver Kupffer cells
Our recent finding that the key molecules involved in iron recycle are highly expressed in BM F4/80+EpoR-eGFP+ macrophages34 suggests the potential role of the EpoR+ macrophages in iron recycle. Given the well-established role of spleen and liver macrophages in the process of iron recycle,36,38 we reasoned that EpoR may be expressed in these tissue macrophages. Figure 6A-a and 6B-a shows that tdTomato was expressed in ∼7% of spleen and ∼23% of liver CD45+F4/80+ macrophages, respectively. Interestingly, in both spleen and liver, tdTomato was only expressed in the CD45+F4/80HighCD11bLow population (Figure 6A-b and 6B-b) but not in the CD45+F4/80-CD11bHigh and CD45+F4/80+CD11bHigh populations (supplemental Figure 9). Spleen red pulp macrophages and liver Kupffer cells are defined as CD45+F4/80HighCD11bLow.37,53 Our findings show that EpoR is expressed in spleen red pulp macrophages and liver Kupffer cells, which are known to be involved in iron recycle.36,38 To show that EpoR is functional, we examined EPO/EpoR signaling in these cells. For this, we enriched macrophages from spleen and liver by using F4/80 beads. The representative cytospin images of the enriched macrophages are shown in supplemental Figure 10A. Flow cytometric analyses revealed that all the enriched macrophages express both F4/80 and tdTomato (supplemental Figure 10B). Western blot analyses show that EPO stimulated Stat5 phosphorylation in the enriched spleen and liver F4/80+tdTomato+ macrophages (Figure 6C-D), indicating EpoR signaling in these cells.
Expression of EpoR-tdTomato in subsets of spleen and liver macrophages, BM HSCs, progenitors, megakaryocytes, and B cells. (A) Flow cytometric analysis of EpoR-tdTomato in spleen macrophages. (a) Left panel: representative plot of F4/80 vs forward scatter height (FSC-H) of CD45+ cells revealing F4/80+ macrophages; right panel: plot of EpoR-tdTomato vs FSC-H of the F4/80+ cells revealing EpoR-tdTomato+ macrophages. (b) Left panel: representative plot of F4/80 vs CD11b of the CD45+ cells; right panel: plot of EpoR-tdTomato vs FSC-H showing the expression of EpoR-tdTomato in the CD11bHigh population. (B) Flow cytometric analysis of EpoR-tdTomato in liver macrophages. (a) Left panel: Representative plot of F4/80 vs FSC-H of CD45+ cells revealing F4/80+ macrophages; right panel: plot of EpoR-tdTomato vs FSC-H of F4/80+ cells revealing the EpoR-tdTomato+ macrophage population. (b) Left panel: Representative plot of F4/80 vs CD11b of CD45+ cells; right panel: plot of EpoR-tdTomato vs FSC-H of F4/80HighCD11bLow showing the expression of EpoR-tdTomato in CD11bHigh population. (C) western blot analysis of pStat5 of the enriched spleen F4/80+ macrophages stimulated with various concentrations of EPO. (D) western blot analysis of pStat5 of the enriched liver F4/80+ macrophages stimulated with various concentrations of EPO. (E) Expression of EpoR-tdTomato in HSCs, MPPs, CMPs, MEPs, GMPs, and CLPs. (F) Left panel: representative plot of CD41 vs FSC-H of CD45+ cells revealing BM megakaryocytes; right panel: plot of EpoR-tdTomato vs FSC-H of CD41+ cells showing the expression of EpoR-tdTomato in subsets of megakaryocytes. (G) Left panel: representative plot of CD19 vs FSC-H of CD45+ cells; right panel: plot of EpoR-tdTomato vs FSC-H of B cells showing the expression of EpoR-tdTomato in subsets of B cells. (H) western blot analysis of pStat5 of the sorted BM CD19+EpoR-tdTomato+ cells stimulated with various concentrations of EPO. N = 3. WT, wild type.
Expression of EpoR-tdTomato in subsets of spleen and liver macrophages, BM HSCs, progenitors, megakaryocytes, and B cells. (A) Flow cytometric analysis of EpoR-tdTomato in spleen macrophages. (a) Left panel: representative plot of F4/80 vs forward scatter height (FSC-H) of CD45+ cells revealing F4/80+ macrophages; right panel: plot of EpoR-tdTomato vs FSC-H of the F4/80+ cells revealing EpoR-tdTomato+ macrophages. (b) Left panel: representative plot of F4/80 vs CD11b of the CD45+ cells; right panel: plot of EpoR-tdTomato vs FSC-H showing the expression of EpoR-tdTomato in the CD11bHigh population. (B) Flow cytometric analysis of EpoR-tdTomato in liver macrophages. (a) Left panel: Representative plot of F4/80 vs FSC-H of CD45+ cells revealing F4/80+ macrophages; right panel: plot of EpoR-tdTomato vs FSC-H of F4/80+ cells revealing the EpoR-tdTomato+ macrophage population. (b) Left panel: Representative plot of F4/80 vs CD11b of CD45+ cells; right panel: plot of EpoR-tdTomato vs FSC-H of F4/80HighCD11bLow showing the expression of EpoR-tdTomato in CD11bHigh population. (C) western blot analysis of pStat5 of the enriched spleen F4/80+ macrophages stimulated with various concentrations of EPO. (D) western blot analysis of pStat5 of the enriched liver F4/80+ macrophages stimulated with various concentrations of EPO. (E) Expression of EpoR-tdTomato in HSCs, MPPs, CMPs, MEPs, GMPs, and CLPs. (F) Left panel: representative plot of CD41 vs FSC-H of CD45+ cells revealing BM megakaryocytes; right panel: plot of EpoR-tdTomato vs FSC-H of CD41+ cells showing the expression of EpoR-tdTomato in subsets of megakaryocytes. (G) Left panel: representative plot of CD19 vs FSC-H of CD45+ cells; right panel: plot of EpoR-tdTomato vs FSC-H of B cells showing the expression of EpoR-tdTomato in subsets of B cells. (H) western blot analysis of pStat5 of the sorted BM CD19+EpoR-tdTomato+ cells stimulated with various concentrations of EPO. N = 3. WT, wild type.
EpoR is expressed in subsets of HSCs, progenitors, megakaryocytes, and B cells
Previous studies have suggested EpoR expression in multipotent hematopoietic cells,7,8 megakaryocytes,9 and B cells.10 However, use of the EpoR-eGFPcre mice failed to detect EpoR expression in any hematopoietic cells.24 Using EpoR-tdTomato-Cre mice, we show here that EpoR-tdTomato was expressed in ∼98% of HSCs, ∼9% of multiple potent progenitors (MPPs), ∼20% of common myeloid progenitors (CMPs), and 97% of megakaryocyte erythrocyte progenitors (MEPs) but not in granulocyte-macrophage progenitors (GMPs) or common lymphoid progenitors (CLPs) (Figure 6E). The hematopoietic hierarchy and gating strategies for HSC and various progenitors are shown in supplemental Figure 11. EpoR-tdTomato was expressed in ∼33% of megakaryocytes (Figure 6F) and ∼3% of B cells (Figure 6G). Moreover, EPO stimulated phosphorylation of Stat5 in the sorted CD19+tdTomato+ cells (Figure 6H). These results show wide expression of EpoR in hematopoietic cells.
EPO leads to changes in EpoR-expressing cells in vivo
We next examined the effects of EPO on EpoR-expressing hematopoietic cells in vivo. EPO injection was administered as previously described.34,54 The scheme of the EPO dosing schedule is shown in supplemental Figure 12A. Flow cytometric analyses of HSCs and various progenitors are shown in supplemental Figure 12B. As positive controls, the effects of EPO injection on erythroid progenitors and erythroblasts were examined. As expected, EPO injection led to a significant increase in CFU-E (supplemental Figure 12C) and erythroblasts (supplemental Figure 12D). Quantification of cell numbers and their proportions in BM is summarized in Table 1, which shows that all EpoR-tdTomato–expressing cells were increased upon EPO injection. In contrast, CLPs and GMPs, which do not express EpoR, were decreased. Furthermore, although EPO injection led to increases in the percentage of CD41+ megakaryocytes within CD45+ cells (supplemental Figure 12E), it led to decreases in the percentage of CD19+ B cells (supplemental Figure 12F). Together, these findings indicate that EPO induces changes biased toward EpoR-tdTomato–expressing cells and induces erythroid lineage bias at all lineage bifurcations during hematopoiesis (supplemental Figure 13).
Effect of EPO injection on hematopoietic stem/progenitor cells, erythroid progenitors, and erythroblasts
| Proportion in BM . | No. in BM . | ||||||
|---|---|---|---|---|---|---|---|
| Cell type . | PBS . | EPO . | P . | Cell type . | PBS . | EPO . | P . |
| HSC (/106) | 3.55 ± 0.48 | 6.02 ± 0.73 | .02541 | HSC (×102) | 4.57 ± 0.52 | 9.16 ± 1.40 | .02200 |
| EpoR-tdTomato+ MPP (/106) | 3.66 ± 0.88 | 13.66 ± 3.07 | .02137 | EpoR-tdTomato+ MPP (×103) | 0.47 ± 0.12 | 1.92 ± 0.38 | .01200 |
| EpoR-tdTomato– MPP (/106) | 33.07 ± 6.99 | 95.25 ± 20.41 | .02857 | EpoR-tdTomato– MPP (×103) | 4.28 ± 0.93 | 13.55 ± 2.53 | .01513 |
| EpoR-tdTomato+ CMP (/103) | 0.05 ± 0.01 | 0.12 ± 0.02 | .00984 | EpoR-tdTomato+ CMP (×104) | 0.86 ± 0.03 | 1.934 ± 0.20 | .00132 |
| EpoR-tdTomato– CMP (/103) | 0.22 ± 0.05 | 0.36 ± 0.04 | .03540 | EpoR-tdTomato– CMP (×104) | 3.68 ± 0.29 | 5.92 ± 0.49 | .00191 |
| GMP (/103) | 0.54 ± 0.08 | 0.26 ± 0.03 | .00863 | GMP (×104) | 7.56 ± 0.78 | 3.72 ± 0.41 | .00326 |
| MEP (/103) | 0.51 ± 0.09 | 1.34 ± 0.18 | .00433 | MEP (×104) | 6.88 ± 1.32 | 18.73 ± 2.55 | .00370 |
| CLP (/106) | 4.66 ± 0.37 | 2.06 ± 0.10 | .03809 | CLP (×102) | 6.99 ± 1.14 | 2.88 ± 0.78 | .03564 |
| BFU-E (/104) | 0.24 ± 0.03 | 0.12 ± 0.01 | .03541 | BFU-E (×104) | 2.94 ± 0.38 | 1.93 ± 0.27 | .09798 |
| CFU-E (/104) | 0.19 ± 0.03 | 0.88 ± 0.06 | .00062 | CFU-E (×104) | 2.49 ± 0.38 | 13.7 ± 0.12 | .00090 |
| Erythroblasts (/102) | 17.87 ± 0.93 | 33.28 ± 1.99 | .00039 | Erythroblasts (×106) | 23.15 ± 1.59 | 50.07 ± 2.64 | .00010 |
| Proportion in BM . | No. in BM . | ||||||
|---|---|---|---|---|---|---|---|
| Cell type . | PBS . | EPO . | P . | Cell type . | PBS . | EPO . | P . |
| HSC (/106) | 3.55 ± 0.48 | 6.02 ± 0.73 | .02541 | HSC (×102) | 4.57 ± 0.52 | 9.16 ± 1.40 | .02200 |
| EpoR-tdTomato+ MPP (/106) | 3.66 ± 0.88 | 13.66 ± 3.07 | .02137 | EpoR-tdTomato+ MPP (×103) | 0.47 ± 0.12 | 1.92 ± 0.38 | .01200 |
| EpoR-tdTomato– MPP (/106) | 33.07 ± 6.99 | 95.25 ± 20.41 | .02857 | EpoR-tdTomato– MPP (×103) | 4.28 ± 0.93 | 13.55 ± 2.53 | .01513 |
| EpoR-tdTomato+ CMP (/103) | 0.05 ± 0.01 | 0.12 ± 0.02 | .00984 | EpoR-tdTomato+ CMP (×104) | 0.86 ± 0.03 | 1.934 ± 0.20 | .00132 |
| EpoR-tdTomato– CMP (/103) | 0.22 ± 0.05 | 0.36 ± 0.04 | .03540 | EpoR-tdTomato– CMP (×104) | 3.68 ± 0.29 | 5.92 ± 0.49 | .00191 |
| GMP (/103) | 0.54 ± 0.08 | 0.26 ± 0.03 | .00863 | GMP (×104) | 7.56 ± 0.78 | 3.72 ± 0.41 | .00326 |
| MEP (/103) | 0.51 ± 0.09 | 1.34 ± 0.18 | .00433 | MEP (×104) | 6.88 ± 1.32 | 18.73 ± 2.55 | .00370 |
| CLP (/106) | 4.66 ± 0.37 | 2.06 ± 0.10 | .03809 | CLP (×102) | 6.99 ± 1.14 | 2.88 ± 0.78 | .03564 |
| BFU-E (/104) | 0.24 ± 0.03 | 0.12 ± 0.01 | .03541 | BFU-E (×104) | 2.94 ± 0.38 | 1.93 ± 0.27 | .09798 |
| CFU-E (/104) | 0.19 ± 0.03 | 0.88 ± 0.06 | .00062 | CFU-E (×104) | 2.49 ± 0.38 | 13.7 ± 0.12 | .00090 |
| Erythroblasts (/102) | 17.87 ± 0.93 | 33.28 ± 1.99 | .00039 | Erythroblasts (×106) | 23.15 ± 1.59 | 50.07 ± 2.64 | .00010 |
Multicolor staining of HSCs and various progenitors were performed by using the enriched lineage– cells. The proportion of HSCs and various progenitor cells in BM was calculated by: proportion of the enriched lineage– cells in total BM cells × proportion of the target cell cluster in the lineage– cells. The numbers of each cell population were calculated by: proportion in BM cells × total BM cells. N = 3. PBS, phosphate-buffered saline.
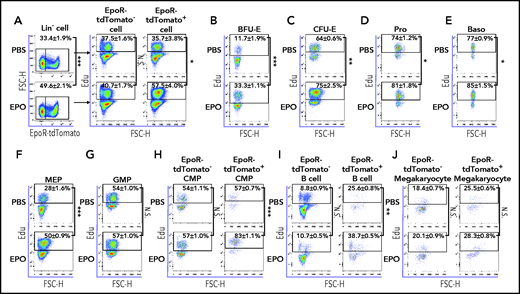
EPO promotes proliferation of the EpoR-expressing cells in vivo
To investigate the mechanisms for EPO-induced increases in EpoR-expressing cells in vivo, we next examined the effect of EPO on cell proliferation using an in vivo Edu incorporation assay.55 As expected, EPO injection led to an increase in the percentage of EpoR+ cells within the Lin– cells (Figure 7A, left panels). Importantly, EPO injection led to a significant increase in the Edu+ percentage of EpoR+ cells but not EpoR– cells (Figure 7A, right panels). Further analyses of various cell types show that the percentages of Edu+ BFU-E (Figure 7B), CFU-E (Figure 7C), proerythroblasts (Figure 7D), basophilic erythroblasts (Figure 7E), and MEPs (Figure 7F) were increased upon EPO injection. In contrast, EPO injection had no effect on Edu uptake by GMPs (Figure 7G), which do not express EpoR. Within CMPs, EPO injection led to an increase in the Edu+ percentage of EpoR+ CMPs but not EpoR– CMPs (Figure 7H). Similarly, EPO injection led to an increase in the Edu+ percentage of EpoR+ but not EpoR– B cells (Figure 7I). Unexpectedly, EPO had no effects on Edu uptake either on EpoR+ or EpoR– megakaryocytes (Figure 7J). Unfortunately, due to the rarity of HSC, MPP, and CLP populations in conjunction with the complexity of multicolor staining as well as fixation and permeabilization, we were unable to detect Edu uptake by these populations. Nevertheless, our findings show that EPO selectively promotes proliferation of the EpoR-expressing cells.
EPO promotes proliferation of EpoR-expressing cells in vivo. (A) Left panels: flow cytometric analyses showing the proportion of EpoR-tdTomato+ and EpoR-tdTomato– populations within lineage– cells with phosphate-buffered saline (PBS) or EPO injection; right panels: flow cytometric analyses showing Edu+ cells of EpoR-tdTomato+ and EpoR-tdTomato– cells with PBS or EPO injection. Flow cytometric analyses displaying proportion of Edu+ cells in BFU-E (B), CFU-E (C), Pro (D), Baso (E), MEP (F), GMP (G), CMP (H), CD19+ B cells (I), and CD41+ megakaryocytes (J) with PBS or EPO injection. N = 3. *P < .05; **P < .01; ***P < .001. FSC-H, forward scatter height; N.S., not significant.
EPO promotes proliferation of EpoR-expressing cells in vivo. (A) Left panels: flow cytometric analyses showing the proportion of EpoR-tdTomato+ and EpoR-tdTomato– populations within lineage– cells with phosphate-buffered saline (PBS) or EPO injection; right panels: flow cytometric analyses showing Edu+ cells of EpoR-tdTomato+ and EpoR-tdTomato– cells with PBS or EPO injection. Flow cytometric analyses displaying proportion of Edu+ cells in BFU-E (B), CFU-E (C), Pro (D), Baso (E), MEP (F), GMP (G), CMP (H), CD19+ B cells (I), and CD41+ megakaryocytes (J) with PBS or EPO injection. N = 3. *P < .05; **P < .01; ***P < .001. FSC-H, forward scatter height; N.S., not significant.
Discussion
EPO is the required cytokine for erythropoiesis, and the expression of its receptor EpoR was originally thought to be restricted in erythroid lineage. Notably, functional studies have shown that EPO plays important roles well beyond erythropoiesis. These include angiogenesis,13,14 neuroprotective activity,15 brain development,21,22 heart development,56-58 regulation of apoptotic cell clearance by macrophages,12 B-cell proliferation and antibody production,10,11,39 megakaryocyte maturation,9 regulation of energy homeostasis,5 and others.59-62 Although the expression of EpoR in non-erythroid cells has been strongly suggested by the functional studies, whether EpoR is expressed in non-erythroid cells has been controversial. In the current study, using the EpoR-tdTomato-Cre mouse model, we readily detected the expression of EpoR-tdTomato in subsets of hematopoietic cells that failed to be detected by using EpoR-eGFPcre mice.24 Our findings have addressed the long-standing controversy on expression of EpoR in hematopoietic cells.
EpoR-eGFPcre mice have been widely used to conditionally delete gene expression, assumed to be selectively in erythroid cells.41-50 Our finding that EpoR was also expressed in other hematopoietic cells challenges this assumption. Moreover, our finding that the EpoR-eGFPcre mice themselves were haploinsufficient raises the concern for use of the EpoR-eGFPcre mouse line for functional studies. These findings suggest that the interpretations of some previous studies using the EpoR-eGFPcre mice as a tool to study the gene function in erythropoiesis may need to be revisited. Particularly, if a gene is expressed in both erythroid cells and EBI macrophages, it will be important to consider whether the altered erythropoiesis is due to changes in erythroid cells, or EBI macrophages, or both cell types.
We recently documented that EBI macrophages are characterized by the expression of EpoR.34 This may seem contradictory to a study by Wang et al,54 the title of which may have given readers the impression that EBI macrophages do not expression EpoR. However, it is important to clarify that the authors did not draw such a conclusion. Instead, based on the finding that EpoR-eGFP was expressed in only ∼5% of BM macrophages,54 it was stated clearly that we did not identify EpoR expression in most of the macrophages and that we still cannot exclude, however, the possibility that a subpopulation of macrophages (the central macrophages binding to erythroblasts) express EpoR on their cell surface.54
Several new findings from the current study further help to clarify this issue. These findings include that EpoR-tdTomato was readily detected in subsets of BM and FL macrophages, the native EBIs were predominantly formed by the F4/80+EpoR-tdTomato+ macrophages, EpoR-Cre led to cre-mediated recombination, and EPO stimulation led to Stat5 phosphorylation in the F4/80+EpoR-tdTomato+ macrophages. A very recent finding from the Paulson laboratory that EpoR signaling in macrophages alters the splenic niche to promote stress erythroid differentiation also supports the notion that EBI macrophages express EpoR.63 Furthermore, the findings that EpoR is expressed in spleen red pulp macrophages and liver Kupffer cells suggest that EPO may enhance iron recycle by red pulp macrophages and Kupffer cells.
It was previously reported that high levels of EPO guide multipotent hematopoietic progenitor cells toward an erythroid fate.8 However, the cellular basis for this effect was not clear. Our findings that EpoR is expressed in HSCs, MPPs, CMPs, and MEPs, that EPO injection led to increases in these EpoR-expressing cells accompanied by decreases in CLPs and GMPs, and that EPO selectively promoted proliferation of the EpoR-expressing cells strongly suggest that high levels of EPO induce erythroid lineage bias at all lineage bifurcations during hematopoiesis via its direct effects on the EpoR-expressing HSCs and progenitor cells. It should be noted that we could not exclude the possibility that the effects of our EPO dosing regimen on HSCs and progenitors are likely to be the consequence of several factors operating during a situation of stress-induced erythropoiesis.
In summary, we generated EpoR-tdTomato-Cre mice that have fully functional EpoR and that enabled identification of EpoR expression in cells that failed to be detected by using the EpoR-eGFPcre mice. The findings that EpoR is expressed in various hematopoietic cells and that EPO injection leads to changes in the EpoR-expressing cells in vivo indicate broad roles of EPO/EpoR in hematopoiesis, which warrant further investigation. We expect that the EpoR-tdTomato-Cre mice will facilitate future studies in the identification of EpoR expression and gene manipulation in many other non-erythroid cells. These studies will ultimately lead to much improved understanding regarding the biology of the EPO/EpoR system.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health, National Heart, Lung, and Blood Institute (HL140625 and HL149626), the Natural Science Foundation of China (U1804282, 81530005, 81570099, and 81770112), and the Rose M. Badgeley Residuary Charitable Trust.
Authorship
Contribution: H.Z., S.W., and D.L. designed experiments, performed research, analyzed the data, and drafted materials and methods; Y.H., C.G., X.G., X.Q., W.L., S.Z., J.G., and L.Z. performed research; A.M., K.Y., and L.C. analyzed the data and edited the manuscript; and X.A. designed experiments, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Xiuli An, New York Blood Center, 310 E 67th St, New York, NY 10065; e-mail: xan@nybc.org.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
REFERENCES
Author notes
H.Z., S.W., and D.L. contributed equally to this work.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal