In this issue of Blood, Casto et al1 describe that the G allele for rs1045642 in the ABCB1 gene in donors reduces the risk of cytomegalovirus (CMV) reactivation after allogeneic hematopoietic stem cell transplantation (HCT) by 20%.2
Human CMV, a ubiquitous β herpesvirus, interacts with target cells during acute and latent infection. CMV infection and disease remains even today one of the major life-threatening infectious complications following allogeneic HCT, despite improvements in diagnostic technologies and prophylactic agents, which have helped to reduce the incidence of CMV-related complications.
Strategies for preventing CMV reactivation and disease after allogeneic stem cell transplantation include antiviral prophylaxis, especially with letermovir, preemptive therapy with (val)ganciclovir and foscarnet, and, in clinical trials, the use of adoptive cellular therapy.3 These strategies are all labor- and cost-intensive, thus tailoring the approaches to allograft recipients at greatest risk and reducing or even omitting prophylaxis and surveillance to the patients at lowest risk are very attractive.
Therefore, clinical scoring systems based on known clinical risk factors (including recipient and donor CMV seropositivity, presence and severity of graft-versus-host disease, and in vivo or in vitro T-cell depletion) have been suggested for better risk stratification for CMV infection and disease among patients undergoing allogeneic stem cell transplantation.4 In addition, reduced intensity conditioning5 and the use of cord blood grafts have also been found to be associated with a higher risk for CMV reactivation and disease.
Genetic variants in both the donor and the recipient have been investigated for an association with CMV reactivation and disease in patients after HCT. Single-nucleotide polymorphisms (SNPs) and other genetic variants may influence the rate and regulatory dynamics of gene transcription, the stability of the messenger RNAs , and the resulting production of biologically active proteins.6 Genetic variants have been found to be associated with susceptibility to inflammation, autoimmune disorders, and increased risk of infections. Particularly genetic variants in genes coding for cytokines, chemokines and cytokine and chemokine receptors have been associated with an increased risk of reactivation and viral diseases.7,8
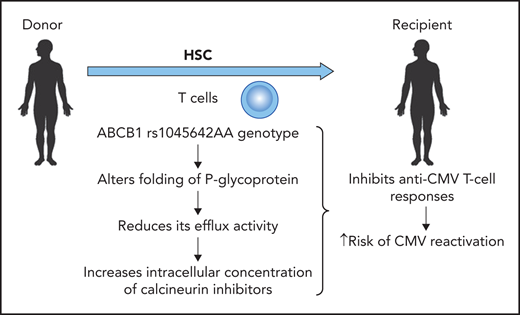
A genetic variant in the ABCB1 gene (rs1045642 AA) in the donor results in an increased risk of CMV reactivation. Professional illustration by Patrick Lane, ScEYEnce Studios.
A genetic variant in the ABCB1 gene (rs1045642 AA) in the donor results in an increased risk of CMV reactivation. Professional illustration by Patrick Lane, ScEYEnce Studios.
In the previous studies, most of the genetic variants reported to be associated with CMV reactivation or disease were identified in only 1 analysis, whereas only a few have been detected in multiple studies. Thus, Casto et al conducted a candidate variant analysis including single-nucleotide and insertion/deletion polymorphisms associated with CMV reactivation and disease to replicate the results of the previous analyses in a large cohort of recipients of an allogeneic stem cell transplantation for which genomic data from both donors and recipients were available. They assessed a cohort of 2169 CMV-seropositive recipients of an allogeneic stem cell graft for 117 candidate variants previously associated with CMV-related phenotypes and in addition carried out a genome-wide association study (GWAS) for CMV reactivation and disease in the same cohort. A GWAS database of this size is unusual especially with well-characterized CMV endpoints and adjustment factors.
Casto et al demonstrated that most genomic variants previously described to be associated with CMV phenotypes do not significantly alter the risk of CMV reactivation and disease. This study represents a semidefinitive assessment of genetics of CMV reactivation in HCT recipients, and the overall result is that the role of genetics is limited, the hazard ratios are not very impressive, and the practical relevance therefore is limited. Thus, overall, this paper reports important negative information on most SNPs and is therefore a valuable contribution to the literature.
However, Casto et al also demonstrated that a variant in the gene encoding P-glycoprotein (rs1045642 A allele) influenced the risk for CMV reactivation (see figure). This genetic variant alters the folding of P-glycoprotein and thus reduces the efflux activity, leading to a higher intracellular concentration of calcineurin inhibitors (cyclosporine, tacrolimus) independent of the whole-blood concentration of these immunosuppressive agents. The higher intracellular concentration of calcineurin inhibitors has a clear impact on lymphocyte function and thus antiviral cellular immunity. The authors describe that the G allele for rs1045642 in the ABCB1 gene in donors reduces the risk of CMV reactivation by 20%, which is comparable to the effect of the donor CMV serostatus in a CMV-seropositive donor. Thus, the magnitude of this genetic variant is similar to what we see with CMV serostatus, which is used to select donors.
Future studies will have to show whether the reduction in therapeutic blood concentration ranges for calcineurin inhibitors in patients receiving their transplant from donors with a homozygous rs1045642 AA genotype will compensate for the lower P-glycoprotein activity and the consecutive higher intracellular concentration of the calcineurin inhibitors cyclosporine and tacrolimus. Potentially, reducing the blood concentration of cyclosporine or tacrolimus in patients with this genetic variant might decrease the risk of CMV reactivation and disease without increasing the risk of acute and chronic GvHD.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal