In this issue of Blood, Pignarre et al characterize the genomic events involved in the cell fate decision between activated B cells and plasmablasts.1 Plasma cells (PCs) play an important role in humoral immunity by synthesizing and secreting antibodies.2 Understanding the biological processes that control the production of PCs is critical to both ensure efficient immune response without autoimmunity or immune deficiency and prevent tumorigenesis. The production of interleukin-4 (IL-4) by follicular helper T cells drives B-cell amplification and maturation.3 However, the full molecular mechanisms behind these functions are not fully understood.
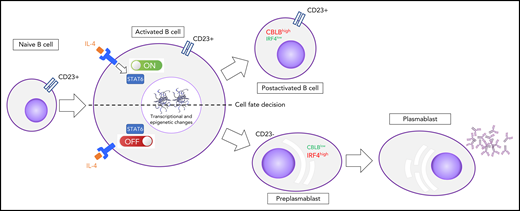
After activation, B cells that are committed to differentiate into PCs downregulate the CD23 cell-surface protein, IL-4/STAT6 signaling, and CBLB activity concomitantly with IRF4 induction. B cells that maintain the IL-4 signaling will not differentiate.
After activation, B cells that are committed to differentiate into PCs downregulate the CD23 cell-surface protein, IL-4/STAT6 signaling, and CBLB activity concomitantly with IRF4 induction. B cells that maintain the IL-4 signaling will not differentiate.
Pignarre et al report new biological events driving normal B- to plasma-cell differentiation. Using an in vitro model, naive B cells were cultured in a 2-step process, which results in differentiation into plasmablasts,4 and the authors demonstrated that cells are destined to differentiate into PCs if there is an early response to IL-4, which results in downregulation of the CD23 cell-surface protein and IL-4/STAT6 signaling. However, B cells maintaining IL-4 signaling did not differentiate. Furthermore, the differentiation of CD23−cells is associated with CBLB E3 ubiquitin ligase downregulation, coinciding with IRF4 induction and with specific chromatin and transcriptional modifications (see figure). The changes were identified by ATAC sequencing and hydroxymethylation profiling. However, no major changes in expression of epigenetic factors were noted. CBLB is known to prevent premature germinal center (GC) exit promoting IRF4 degradation in light zone B cells.5 Pignarre et al reported potential STAT6 binding sites in the CBLB promoter, suggesting potential direct regulation, hence the interest in characterizing STAT6 targets, using chromatin immunoprecipitation. CD23− B cells, postactivation, have the characteristics of preplasmablasts with a significant increase in chromatin accessibility at immunoglobulin heavy chain coding loci. Full transcriptomic characterization of the proposed model at a single-cell level would be particularly useful in deciphering the heterogeneity and transcriptional trajectories during B- to plasma-cell differentiation.
The major transcriptional and epigenetic changes reported by Pignarre et al may be associated with changes in nuclear organization during terminal B-cell differentiation. Gene regulation depends on the 3-dimensional chromatin organization and its regulatory elements. Recent data obtained in a mouse model revealed that B- to plasma-cell differentiation is associated with major changes in chromosome topology that could be driven by PC biology or reflect enhancer-induced modifications in chromatin organization.6 B- to plasma-cell transition is associated with compartmentalization changes along with gain in genomic interactions across the Prdm1 locus, increasing genomic interactions between promoter regions and regulatory elements, concurrent with transcriptional induction. In contrast, the early B-cell factor 1 (Ebf1) locus repositions to pericentromeric heterochromatin in association with transcriptional repression. Furthermore, the interchromosomal hubs reported during B- to plasma-cell maturation are associated with histone marks that define transcriptionally active or repressive hubs. The epigenetic landscape characterization together with nuclear architecture study of the human B- to plasma-cell differentiation model developed by Pignarre et al may provide important findings for the understanding of the molecular mechanisms driving B- to plasma-cell fate.
IL-4 production by T-follicular helper and STAT6 mutations activates and drives the IL-4/STAT6 axis in follicular lymphoma.7 Recently, single-cell RNAseq characterization of purified GC B cells (GCBs) generated a new single-cell cell of origin classification that identified distinct prognostic subgroups within the GCB and activated B-cell–like diffuse large B-cell lymphoma subgroups.8 The analysis of these single-cell transcriptomic resources derived from GC purified B cells, in light of the new data provided by Pignarre et al, may be of particular interest. The results provided by Pignarre et al provide new insights in the molecular mechanisms driving follicular lymphoma biology.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal