In this issue of Blood, Jäger et al report that interferon lambda 4 (IFNL4) is an important genetic determinant of interferon-α (IFN-α)–induced molecular response (MR) in myeloproliferative neoplasms (MPNs), allowing a better stratification of patients.1
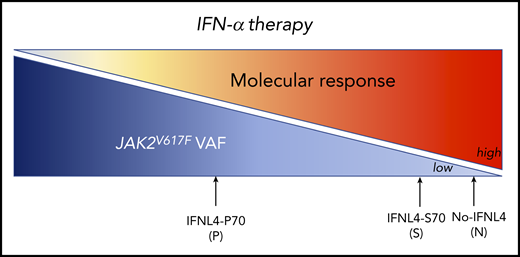
The IFN-α therapy induces an MR on JAK2V617F PV patients according to the IFNL4 functional status: very high MR for N: no IFNL4 and S: IFNL4-S70 and less MR for P: IFNL4-P70.
The IFN-α therapy induces an MR on JAK2V617F PV patients according to the IFNL4 functional status: very high MR for N: no IFNL4 and S: IFNL4-S70 and less MR for P: IFNL4-P70.
BCR-ABL-negative MPNs include polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF). They result from the transformation of a hematopoietic stem cell leading to the overproduction of mature cells (red blood cells, platelets, granulocytes). The 3 main drivers are JAK2V617F in all three of the diseases, and mutations in the thrombopoietin receptor (MPL) and calreticulin (CALR) genes only in ET and PMF. They are generally treated with drugs that normalize the hematological parameters, ameliorate the symptoms, and decrease the splenomegaly, but have minor impact on eliminating JAK2V617F or CALR-mutated cells. Type I IFN-α has emerged as an interesting therapy because it induces both a hematologic response (HR) in >70% of ET/PV patients and an MR in >50% of patients.2,3 However, there is considerable room for improvement as only 20% of patients achieved a major MR response with a remission after treatment. In addition, long-term IFN-α therapy may induce significant side effects (flulike symptoms, tiredness, depression, etc), and ∼10% of patients are intolerant of this therapy. Therefore, it is important either to improve our understanding of its mechanism of action to develop more effective therapy or to predict the response to IFN-α therapy.
Jäger et al have investigated the implication of germline genetic factors in the HR and MR in MPNs using a cohort of one hundred PV patients treated with ropeg-IFN-α-2b for 36 months (PROUD-PV and CONTINUATION-PV clinical trials). First, they performed a global unbiased genomic approach (genome-wide association studies [GWASs] analysis), but found no allelic association with IFN-α–induced HR or MR in contrast to what was reported in IFN-α therapy of hepatitis C. Because statistical power of GWASs is low with a cohort of relatively limited size compared with much larger cohorts of hepatitis patients, the authors focused on the IFNL4 variants targeted analysis. The IFNL4 gene encodes the type III IFN-λ4, and several variants were reported to be highly involved in the hepatitis C virus (HCV) clearance and to correlate with an efficient IFN-α therapy.4
Jäger et al provide evidence that 2 intronic IFNL4 variants (rs8099917 and rs12979860) that have been previously identified as impacting IFN-α–induced HR in ET/PV patients5 also had a positive effect on IFN-α–induced MR after 36 months of therapy. Moreover, they analyzed the effect on MR of 2 other exonic IFNL4 variants never investigated in MPNs as individual or combined (haplotype pair) genotypes (rs368234815 and/or rs117648444). Although the rs368234815_ΔG results in a normal full-length protein (IFNL4-P70), the rs368234815_TT does not produce any protein (no-IFNL4) due to a premature stop codon. Moreover, the rs117648444 [G>A] leads to a switch of proline to serine, creating an IFNL4-S70 protein with impaired activity compared with the IFNL4-P70 protein.6 In individual analysis, the authors observed that harboring no-IFNL4 had a significant positive impact on the rate of MR response. The patients were then grouped into 3 main functional categories: patients not producing any functional IFNL4 (no-IFNL4), producers of the impaired IFNL4-S70, and producers of the fully functional IFNL4-P70. Although the combined analyses based on IFNL4 functionality did not reveal any associations with HR, for MR, a strong difference in response rates between the patients with no-IFNL4 (89.3% responders) and IFNL4-P70 (43.3% responders) was observed. The effect of the impaired variant IFNL4-S70 resembled the category with no-IFNL4. Altogether, these results indicate that IFN-λ4 full-length protein negatively impacts the IFN-α–induced MR (see figure). However, it remains to be determined if these genotypes behave similarly in CALR- and MPL-mutated patients using cohorts of ET. Interestingly, as the JAK2V617F variant allele frequency (VAF) is extremely variable in MPNs depending on the disease and even among patients with the same disease, the authors explored whether the baseline VAF correlates with the presence of the IFNL4 exonic variants. Indeed, in HCV, the spontaneous remission strongly correlates with the absence of IFNL4.7 The IFNL4 functional status did not influence the baseline JAK2V617F VAF, but it did impact the MR independent of baseline VAF. In addition, it will be interesting to investigate in larger cohorts whether some genotypes are underrepresented in MPNs compared with control cohorts to explore the role of IFNL4 in disease development.
The most important question is how the negative impact of IFN-λ4 on IFN-α MR occurs so that IFN-α therapy may be improved. Although these 2 IFNs act through different IFNAR1 and IFNAR2 receptors for IFN-α and IFNLR1 and IL10R2 receptors for IFN-λ4, they can both activate the JAK/STAT signaling pathway in a similar manner. After ligand binding to their respective receptors, the phosphorylation of JAK1 and TYK2 induces the phosphorylation of the receptor leading to the recruitment and phosphorylation of STAT1 and STAT2, which heterotrimerize with IFN-regulatory factor 9 to form the IFN-stimulated gene factor 3.8 This complex migrates into the nucleus to induce the IFN-stimulating genes (ISGs). The IFN-λ4 can interfere with other IFN-α–mediated signaling through an increase in negative regulators, such as SOCS1, ISG15, and USP18, whose binding is very specific for IFNAR2. Indeed, ISG overexpression was predicted to be a deleterious factor for the treatment of HCV with IFN-α therapy.9 Thus, an impaired IFN-λ4 activity (IFNL4-S70) would alleviate the negative regulation of IFN-λ4. Nevertheless, the IFN-α was shown in mouse models to directly target JAK2V617F hematopoietic stem cells through IFNAR receptors.10 Thus, it is difficult to hypothesize that the IFN-λ4 can directly act on hematopoietic stem and progenitor cells because the IL10R2 and IFNLR1 expression seems to be restricted to epithelial liver cells and eventually to immune cells, which alternatively can rather contribute to MR by an indirect immunomodulatory role. Thus, in MPNs, the effects of IFN-α are essentially cell-autonomous, whereas those of IFN-λ4 might be non–cell-autonomous.
In aggregate, the study of Jäger et al importantly points to a favorable IFNL4 genotype/phenotype that should be taken into consideration in the treatment of PV with IFN-α.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal