In this issue of Blood, Ruben et al present detailed structures of coagulation factor V (FV) and FVa, generated through cryogenic electron microscopy (cryo-EM). This study provides, for the first time, structural detail on conformational changes in FV upon its activation, which control not only its ability to enhance coagulation but also how it is regulated and degraded.1
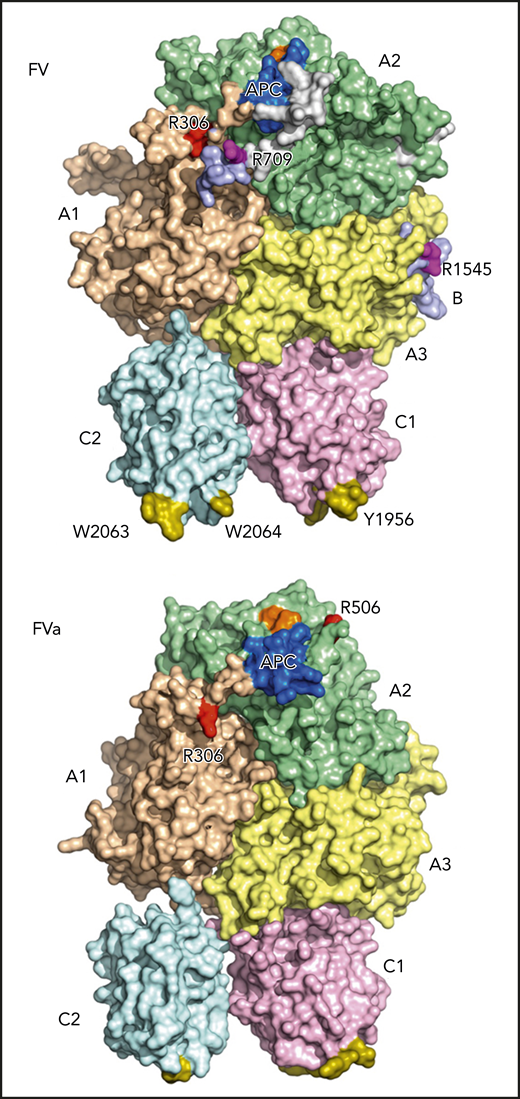
Cryo-EM structures of FV and FVa. The domains of both proteins are shown in wheat (A1), pale green (A2), light blue (B), pale yellow (A3), light pink (C1), and pale cyan (C2). The FV/FVa C domains provide a platform supporting the A domains. The A1 and A3 domains sit on top of the C domains, with the A2 domain resting between the A1 and A3 domains and having no contact with the C domains. The A1-A2-A3-C1-C2 domain assembly is resolved, whereas the B domain is more dynamic and only visible at the most N- and C-terminal segments, directly connected with the A2 and A3 domains, respectively. The sites of thrombin activation at Arg709 and Arg1545 (magenta) are clearly visible in the FV structure. The sites of APC cleavage at Arg306 and Arg506 are largely buried in FV but become more exposed after thrombin-mediated activation (please compare FV and FVa structures), priming FVa for APC-mediated inactivation. The spikes in the C domains responsible for membrane binding are shown in olive, and the FXa- and APC-binding epitopes are shown in orange and blue, respectively. Amino acid residues are highlighted using their 1-letter codes. The figure has been adapted from Figure 2B,E in the article by Ruben et al that begins on page 3137.
Cryo-EM structures of FV and FVa. The domains of both proteins are shown in wheat (A1), pale green (A2), light blue (B), pale yellow (A3), light pink (C1), and pale cyan (C2). The FV/FVa C domains provide a platform supporting the A domains. The A1 and A3 domains sit on top of the C domains, with the A2 domain resting between the A1 and A3 domains and having no contact with the C domains. The A1-A2-A3-C1-C2 domain assembly is resolved, whereas the B domain is more dynamic and only visible at the most N- and C-terminal segments, directly connected with the A2 and A3 domains, respectively. The sites of thrombin activation at Arg709 and Arg1545 (magenta) are clearly visible in the FV structure. The sites of APC cleavage at Arg306 and Arg506 are largely buried in FV but become more exposed after thrombin-mediated activation (please compare FV and FVa structures), priming FVa for APC-mediated inactivation. The spikes in the C domains responsible for membrane binding are shown in olive, and the FXa- and APC-binding epitopes are shown in orange and blue, respectively. Amino acid residues are highlighted using their 1-letter codes. The figure has been adapted from Figure 2B,E in the article by Ruben et al that begins on page 3137.
Coagulation is initiated upon vascular injury as an innate response to prevent blood loss. FV is central to this process by acting as a cofactor for FXa within the prothrombinase complex.2 In the absence of vascular injury, FV circulates as a procofactor comprising an A1-A2-B-A3-C1-C2 domain structure. As a procofactor, FV has no procoagulant functions but instead serves as an anticoagulant regulator by enhancing 2 anticoagulant pathways, the tissue factor pathway inhibitor (TFPI) and the activated protein C (APC) pathways.3 Only once coagulation has been initiated is FV activated to FVa by limited proteolysis at 3 sites, Arg709, Arg1018, and Arg1545, resulting in complete removal of the B domain.2 The procoagulant functions of FVa are in turn regulated by APC-mediated proteolysis at Arg306 and Arg506.2,3 Although we have a general understanding of the molecular mechanisms behind the pro- and anticoagulant functions of FV/FVa, structural information has been lacking. Previous studies have provided some structural insight into FVa or its inactive derivatives.2,4-7 However, these have not included an experimentally determined structure of the human FV A2 domain. This is of particular importance, because this domain is key to both the procoagulant cofactor function of FVa as well as its inactivation by APC.2 Ruben et al have for the first time determined the structures of both FV and FVa, including their A2 domains (see figure).
The structures illustrate how the 2 FV C domains provide a platform supporting the A domains, similar to the previously published structure of bovine FVai.6 The A1 and A3 domains sit on top of the C domains, with the A2 domain resting between the A1 and A3 domains and having no contact with the C domains (see figure). In contrast to the A and C domains, the B domain is overall disordered, looping around the protein in a dynamic conformation. Unfortunately, this means that there is no structural insight into the FV B-domain acidic and basic regions, which are essential in maintaining FV in a procofactor state.2 However, the most N- and C-terminal segments of the B domain, directly connected with the A2 and A3 domains, were more stable, crucially allowing the resolution of the functionally important Arg709 and Arg1545 thrombin cleavage sites.
Through comparing the FV and FVa structures (see figure), the authors show how removal of the B domain from FV resulted in increased disorder in the A domains, mostly in the C-terminus of the A2 domain, as well as (somewhat surprisingly) in the C2 domain (FV/FVa structures are shown from additional angles in Figure 2 of the article by Ruben et al). These changes in conformation and subsequent functional epitope exposure in the A2 domain are most likely necessary for FVa to assemble with FXa in the prothrombinase complex. Furthermore, although the APC Arg306 and Arg506 cleavage sites are largely buried in FV, they become more exposed after thrombin-mediated activation of FV, essentially priming FVa for APC-mediated inactivation. The relatively inefficient cleavage by APC of the Arg506 cleavage site in FV, compared with that in FVa, is therefore explained.8 The large distance between the Arg306 and Arg506 APC cleavage sites supports independent cleavage. Indeed, biochemical studies have shown a significant difference in cleavage kinetics between the 2 sites. Furthermore, their differing dependence for cleavage on protein S, the cofactor of APC, may be partially explained by this spatial separation.3,9
In addition to structural insight into overall FV/FVa function and regulation, this study also suggests explanations for the structural impact of various naturally occurring mutations in the FV gene. The authors discuss how the FV Leiden,9 FV Besanҫon,10 and FV Nara3 thrombophilic mutations are likely to affect the structure of FV/FVa. The latter 2 likely destabilize the C2 and C1 membrane interactions, essential for FV/FVa activation and regulation.
One of the anticoagulant functions of FV is caused by FV interactions with TFPIα through the acidic region in the B domain, which is exposed in partially activated forms of FV as well as in naturally occurring FV splice variants.9 Splice variants of FV, such as the FV short, which is upregulated in East Texas bleeding disorder,9 have a truncated B domain and are single-chain proteins. Following from the present study, it is tempting to speculate that they may be amenable to cryo-EM structural determination with the potential of providing further information on the TFPIα interaction site. Also, as mentioned by the authors, the present structures are of FV/FVa in isolation. Future structural determination of the prothrombinase and FVa inactivation complexes may now be possible and, although a challenge, will provide even greater insight into the mechanisms surrounding the various functions of FV and FVa.
Conflict-of-interest disclosure: The author declares no competing financial interests.