Key Points
Eosinophil differentiation is delayed and reduced in the absence of ATG5 under both physiological and leukemic conditions.
Effector functions of ATG5-deficient mouse and human eosinophils are enhanced under in vivo conditions.
Abstract
Eosinophils are white blood cells that contribute to the regulation of immunity and are involved in the pathogenesis of numerous inflammatory diseases. In contrast to other cells of the immune system, no information is available regarding the role of autophagy in eosinophil differentiation and functions. To study the autophagic pathway in eosinophils, we generated conditional knockout mice in which Atg5 is deleted within the eosinophil lineage only (designated Atg5eoΔ mice). Eosinophilia was provoked by crossbreeding Atg5eoΔ mice with Il5 (IL-5) overexpressing transgenic mice (designated Atg5eoΔIl5tg mice). Deletion of Atg5 in eosinophils resulted in a dramatic reduction in the number of mature eosinophils in blood and an increase of immature eosinophils in the bone marrow. Atg5-knockout eosinophil precursors exhibited reduced proliferation under both in vitro and in vivo conditions but no increased cell death. Moreover, reduced differentiation of eosinophils in the absence of Atg5 was also observed in mouse and human models of chronic eosinophilic leukemia. Atg5-knockout blood eosinophils exhibited augmented levels of degranulation and bacterial killing in vitro. Moreover, in an experimental in vivo model, we observed that Atg5eoΔ mice achieve better clearance of the local and systemic bacterial infection with Citrobacter rodentium. Evidence for increased degranulation of ATG5low-expressing human eosinophils was also obtained in both tissues and blood. Taken together, mouse and human eosinophil hematopoiesis and effector functions are regulated by ATG5, which controls the amplitude of overall antibacterial eosinophil immune responses.
Introduction
Eosinophils are bone marrow–derived granulocytes with versatile effector functions in innate and adaptive immunity.1 The differentiation of eosinophils is a complex and tightly organized process. The first step is a specific lineage commitment of multipotent hematopoietic stem cells. In humans, the common myeloid progenitors (CMPs) progress to eosinophil lineage–committed progenitors,2 whereas murine CMPs give rise to the granulocyte-monocyte progenitors before diverging into eosinophil lineage–committed progenitors.3 The process of differentiation is regulated by transcription factors and cytokines. Several key transcription factors that control eosinophil development have been identified, such as GATA-1,3,4 GATA-2,5 PU.1,6,7 C/EBP,8,9 and XBP1.10 The most important growth factors in promoting eosinopoiesis are interleukin-3 (IL-3), granulocyte-macrophage colony-stimulating factor (GM-CSF), and IL-5, the latter being the most potent and characteristic cytokine for the eosinophil lineage.11
In addition to transcription factors and eosinophil hematopoietins, several studies suggested additional players involved in the regulation of eosinophil hematopoiesis. For instance, RhoH was reported to counter-regulate eosinophil differentiation and maturation, likely by regulating GATA-2 levels and the cell cycle.12 An inhibitory effect on eosinophil differentiation was also shown for interferon-γ,13 IL-17,14 and paired immunoglobulin-like receptor A.15 Conversely, eosinophil differentiation was induced in the presence of protein tyrosine phosphatase SHP2 via the regulation of the extracellular signal–regulated kinase pathway.16
Autophagy is a highly regulated catabolic process in which cellular contents are sequestered in double-membrane vesicles called autophagosomes, followed by their degradation with proteolytic lysosomal enzymes.17 Autophagy-related protein 5 (ATG5) is a key component of the autophagic pathway18 and has been implicated in the regulation of both innate and adaptive immunity. For example, ATG5 was shown to regulate the differentiation of B cells,19 plasma cells,20 neutrophils,21,22 and innate lymphoid cells.23 ATG5 is also critical for cytokine secretion,24 elimination of pathogens,25,26 and antigen presentation.27,28 However, the importance of ATG5 and autophagy for eosinophil differentiation and function remains unknown.29 Addressing this question seems to be important because ATGs have been implicated in the pathogenesis of eosinophilic diseases such as asthma and eosinophilic esophagitis (EoE).30,31 Moreover, autophagy has recently been implicated in the formation of eosinophil extracellular traps (EETs).32 Therefore, investigating the role of ATGs and autophagy in the eosinophil lineage is an unmet scientific need with medical implications.
The current study investigated the impact of ATG5 on eosinophil hematopoiesis and effector functions. We used a genetic approach to generate mice specifically lacking the Atg5 gene within the eosinophil lineage and also analyzed human eosinophils under pathologic conditions. We show the involvement of ATG5 in the regulation of eosinophil differentiation in both mouse and human systems. Moreover, Atg5-knockout mouse eosinophils and ATG5low-expressing human eosinophils exhibit enhanced degranulation abilities, suggesting that eosinophil effector functions are influenced by autophagy.
Material and methods
Atg5eoΔ and Atg5eoΔII5tg mice
Animal studies were approved by the Veterinary Office of the Canton of Bern and conducted in accordance with Swiss federal legislation on animal welfare under animal license number 49/18. Atg5flox/floxeoCre mice (designated Atg5eoΔ) were generated by crossing Atg5flox/flox mice33 (Atg5tm1Myok) with eoCre mice34 (Epxtm1.1(cre)Jlee), as previously described.35 Atg5flox/flox mice were kindly provided by C. Münz (University of Zürich, Zurich, Switzerland). Atg5flox/floxeoCreIl5tg mice (designated Atg5eoΔIl5tg) were generated by crossing Atg5eoΔ mice with Il5tg mice36 [Tg(Cd3d-Il5)NJ.1638Nal] to increase the number of eosinophils for functional in vitro assays. EoCre and Il5tg mice were kindly provided by J. J. Lee (Mayo Clinic, Phoenix, AZ). EoCre (designated control) and eoCreIl5tg mice (designated CtrlIl5tg) were used as controls in all experiments.
Other detailed methods
All other methods are described in the supplemental Information (available on the Blood Web site).
Results
Knocking out Atg5 in the eosinophil lineage results in delayed and reduced eosinophil precursor proliferation and maturation
Due to observed neonatal lethality in systemic Atg5 gene knockout mice,37 conditional tissue-specific Atg5-deficient mice (Atg5flox/flox mice)33 have been generated to elucidate the functions of ATG5. In this study, we crossed Atg5flox/flox mice with a knock-in strain of mice expressing Cre recombinase under the control of the eosinophil peroxidase (EPX) promoter, which is exclusively active in eosinophils (eoCre mice).34 Cre recombinase expression in eoCre mice occurs with no evidence of cytotoxicity in the eosinophil lineage–committed progenitors, which are the earliest identifiable cells in the eosinophil lineage. Generated Atg5flox/floxeoCre mice (Atg5eoΔ mice) were used to investigate the effects of Atg5 deletion within the eosinophil lineage.34 In all experiments, heterozygous eoCre mice (eoCre+/−) were used because they expressed sufficient amounts of Cre recombinase to excise the targeted gene while still maintaining half of the total EPX protein levels.34
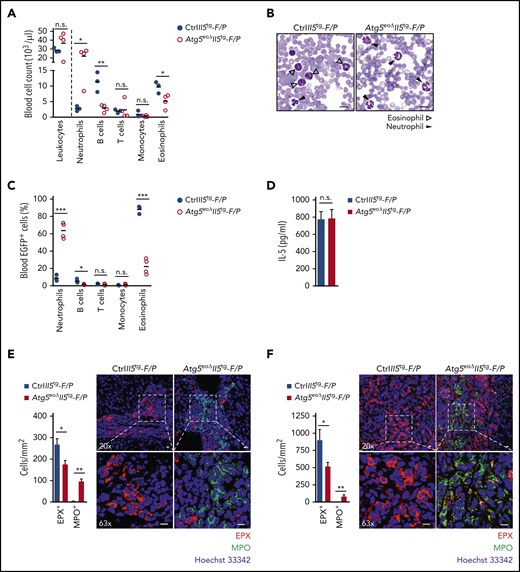
To examine the role of Atg5 in the bone marrow eosinophils, we analyzed their maturity by measuring the cell surface expression of the chemokine receptor CCR3, which is expressed on fully mature eosinophils.38 Eosinophils were identified as SSChi, Siglec-F+, and Gr-1lo-int cells (supplemental Figure 1A). Elevated relative (Figure 1A) and absolute (Figure 1B) numbers of immature eosinophils (ie, Siglec-F+, CCR3–) in the bone marrow of Atg5eoΔ mice were observed compared with control mice, suggesting a role for Atg5 in eosinophil differentiation. Reduced eosinophil differentiation has been reported in mice lacking IL-5Rα or IL-33R (ST2).39,40 We observed slightly reduced expression of IL-5Rα on bone marrow–derived Atg5-knockout eosinophils (supplemental Figure 1B), whereas no difference was observed regarding IL-33R expression (supplemental Figure 1C). In the circulation, control and Atg5-knockout eosinophils appeared as a homogenous mature population (ie, Siglec-F+, CCR3+), indicating that only mature eosinophils exit the bone marrow regardless of the presence or absence of Atg5 (supplemental Figure 1D). However, blood eosinophil numbers were decreased in Atg5eoΔ mice, again pointing to reduced eosinophil differentiation. No difference was observed in the numbers of blood lymphocytes, monocytes, and neutrophils between Atg5eoΔ and control mice (supplemental Figure 1E).
Knocking out Atg5 in the eosinophil lineage results in delayed and reduced eosinophil precursor proliferation and maturation. (A) Flow cytometry. Maturity of eosinophils in the bone marrow of control and Atg5eoΔ mice was assessed by measuring the cell surface expression of chemokine receptor CCR3. Mature eosinophils were defined as Siglec-F+CCR3+ and immature eosinophils as Siglec-F+CCR3– cell populations (n = 6). Right: representative original flow cytometry data are shown. Each symbol represents a value for an individual mouse throughout. (B) The data of panel A are presented as absolute numbers. (C) Flow cytometry. Eosinophils were differentiated in vitro from bone marrow cells of control and Atg5eoΔ mice, and their maturity was assessed by measuring the surface expression of CCR3 on the indicated days (n = 3). Right: representative original flow cytometry data are shown. (D) Flow cytometry. Proliferating eosinophil precursors in the S phase of the cell cycle during in vitro differentiation were detected by 5-ethynyl-2'-deoxyuridine (EdU) labeling (n = 3). (E) Flow cytometry. The proliferative status of eosinophil precursors in the G1, S, G2, and M phases of the cell cycle during in vitro differentiation was determined by Ki-67 expression measurements (n = 3). (F) [3H]‐thymidine incorporation assay. For each time point, [3H]‐thymidine was added to the cultures of in vitro differentiating eosinophils 24 hours before the measurement. Radioactivity in DNA recovered from the cells was determined with a scintillation counter to determine the extent of cell division (n = 3). (G) CFU assay. Freshly isolated bone marrow cells from control and Atg5eoΔ mice were plated in methylcellulose media supplemented with recombinant mouse IL-5. After 12 days of incubation, cells were stained with the substrate for EPX, and brown-stained Eo-CFUs of >50 cells were counted in each well (n = 5). Right: representative images of culture wells and single colonies are shown. Scale bars, 10 µm. To confirm the identification of colonies as Eo-CFU, samples were selected and removed from the wells for analysis of Siglec-F expression by flow cytometry as presented in supplemental Figure 1H. (H) Immunoblotting. Protein lysates during in vitro culture of differentiating eosinophils were collected on the indicated days and analyzed for the presence of phosphorylated p38 (Thr180/Tyr182), phosphorylated p44/42 (Thr202/Tyr204), ATG5, and LC3 protein expression. p38, p44/42, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) protein levels were analyzed as loading controls. Representative immunoblots of 3 independent experiments are shown. Values are means ± standard error of the mean, or single data are presented in scatter dot plots in which the medians are indicated as red lines. *P < .05; **P < .01; ***P < .001.
Knocking out Atg5 in the eosinophil lineage results in delayed and reduced eosinophil precursor proliferation and maturation. (A) Flow cytometry. Maturity of eosinophils in the bone marrow of control and Atg5eoΔ mice was assessed by measuring the cell surface expression of chemokine receptor CCR3. Mature eosinophils were defined as Siglec-F+CCR3+ and immature eosinophils as Siglec-F+CCR3– cell populations (n = 6). Right: representative original flow cytometry data are shown. Each symbol represents a value for an individual mouse throughout. (B) The data of panel A are presented as absolute numbers. (C) Flow cytometry. Eosinophils were differentiated in vitro from bone marrow cells of control and Atg5eoΔ mice, and their maturity was assessed by measuring the surface expression of CCR3 on the indicated days (n = 3). Right: representative original flow cytometry data are shown. (D) Flow cytometry. Proliferating eosinophil precursors in the S phase of the cell cycle during in vitro differentiation were detected by 5-ethynyl-2'-deoxyuridine (EdU) labeling (n = 3). (E) Flow cytometry. The proliferative status of eosinophil precursors in the G1, S, G2, and M phases of the cell cycle during in vitro differentiation was determined by Ki-67 expression measurements (n = 3). (F) [3H]‐thymidine incorporation assay. For each time point, [3H]‐thymidine was added to the cultures of in vitro differentiating eosinophils 24 hours before the measurement. Radioactivity in DNA recovered from the cells was determined with a scintillation counter to determine the extent of cell division (n = 3). (G) CFU assay. Freshly isolated bone marrow cells from control and Atg5eoΔ mice were plated in methylcellulose media supplemented with recombinant mouse IL-5. After 12 days of incubation, cells were stained with the substrate for EPX, and brown-stained Eo-CFUs of >50 cells were counted in each well (n = 5). Right: representative images of culture wells and single colonies are shown. Scale bars, 10 µm. To confirm the identification of colonies as Eo-CFU, samples were selected and removed from the wells for analysis of Siglec-F expression by flow cytometry as presented in supplemental Figure 1H. (H) Immunoblotting. Protein lysates during in vitro culture of differentiating eosinophils were collected on the indicated days and analyzed for the presence of phosphorylated p38 (Thr180/Tyr182), phosphorylated p44/42 (Thr202/Tyr204), ATG5, and LC3 protein expression. p38, p44/42, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) protein levels were analyzed as loading controls. Representative immunoblots of 3 independent experiments are shown. Values are means ± standard error of the mean, or single data are presented in scatter dot plots in which the medians are indicated as red lines. *P < .05; **P < .01; ***P < .001.
To test the effect of Atg5 on eosinophil hematopoiesis more directly, we used an established in vitro model of eosinophil differentiation from the mouse bone marrow.41 In brief, the bone marrow cells were supplemented with stem cell factor and FLT3-ligand from day 0 to 4. Starting on day 4, the medium was replaced with IL-5–containing medium only, and the cells were cultured for an additional 8 days in the presence of IL-5. We analyzed eosinophil maturation and observed a significantly reduced population of mature eosinophils (ie, Siglec-F+, CCR3+) when Atg5 was absent, indicating that eosinophil differentiation is delayed and reduced under these conditions (Figure 1C). To investigate whether the effect of Atg5 on eosinophil maturation is direct and primarily mediated by ATG5, we re-expressed Atg5 in the bone marrow cells of Atg5eoΔ mice that was followed by a more rapid and efficient in vitro eosinophil differentiation (supplemental Figure 1F).
We also assessed the proliferative status of in vitro differentiating eosinophils by measuring the 5-ethynyl-2'-deoxyuridine labeling and Ki-67 protein expression. In the absence of Atg5, a significant reduction was observed in 5-ethynyl-2'-deoxyuridine–positive (Figure 1D) and Ki-67–positive (Figure 1E) eosinophil precursors compared with control cells. We also investigated cell proliferation using a [3H]-thymidine incorporation assay and again found evidence for decreased proliferation of Atg5-knockout eosinophil precursors at days 11 and 12 after initiating the process of differentiation in vitro (Figure 1F). To examine a possible effect of Atg5 on eosinophil survival, cell viability of eosinophils during in vitro differentiation was measured, and no difference was observed between Atg5-knockout and control eosinophil precursors (supplemental Figure 1G).
To study the proliferation and differentiation pattern of eosinophil progenitors according to their ability to form colonies, the colony-forming unit (CFU) assay was performed. Each colony in a semisolid methylcellulose-based medium represents the progeny of a single eosinophil progenitor,42 defined as IL-5Rα+Lin−Sca-1−CD34+c-Kitlow.3 The numbers of eosinophil CFUs (Eo-CFUs) derived from the bone marrow of Atg5eoΔ mice were reduced and appeared to be diffused and less aggregated compared with control Eo-CFUs (Figure 1G). The cells in colonies had homogeneous morphology, and they were stained with o-phenylenediamine, which is a substrate for EPX. Selected stained colonies were examined for the surface expression of Siglec-F and CCR3. Based on the positive Siglec-F staining (>97%), we concluded that the colonies represent Eo-CFUs (supplemental Figure 1H). Control and Atg5-knockout eosinophils derived from these colonies reached the same high levels of CCR3 (supplemental Figure 1I). These data support the assumption that the Atg5 gene allows an efficient differentiation of the eosinophil lineage, but its absence does not block the maturation process.
Knocking out Atg5 results in reduced activation of p38 and p44/42 MAPKs in eosinophil precursor cells
The MAPK pathway is involved in cell proliferation and differentiation of eosinophil lineage mainly through the activity of p38 MAPK.43 To explore a potential connection between levels of ATG5 and MAPK signaling pathways, we analyzed the extent of p38 and p44/42 phosphorylation during in vitro differentiation of Atg5-knockout and control eosinophils. In Atg5-knockout eosinophils, decreased levels of phosphorylated p38 were detected on days 11 and 12 during in vitro differentiation, together with decreased levels of phosphorylated p44/42 on days 10, 11, and 12 (Figure 1H). As expected, eosinophils differentiated in vitro from the bone marrow of Atg5eoΔ mice did not express ATG5 (detected as an ATG5-ATG12 conjugate),44,45 resulting in a decreased conversion of cytosolic LC3-I to the autophagosomal marker LC3-II.46 These data confirmed that autophagy is suppressed in Atg5-knockout eosinophils.
We next used pharmacologic inhibitors of p38 (PD169316 and SB203580) and p44/42 (PD98059 and U0126) MAPK pathways in a concentration-dependent manner to investigate their effect on eosinopoiesis.21,43,47 In the course of eosinophil differentiation from the bone marrow cells of control mice, PD169316 and SB203580 efficiently inhibited the maturation (supplemental Figure 2A-B, upper panels) along with the proliferation of eosinophil precursors (supplemental Figure 2A-B, middle panels). In contrast, PD98059 and U0126 had no effect on eosinophil maturation (supplemental Figure 2C-D, upper panels) but were able to reduce cell proliferation (supplemental Figure 2C-D, middle panels). Other than PD98059 at the highest concentration (10 µM), no other inhibitor induced cell death at the designated concentrations (supplemental Figure 2A-D, lower panels). Taken together, decreased differentiation of eosinophils in the absence of Atg5 could be explained by reduced MAPK activities.
Because eosinophils in mice comprise only up to 5% of leukocytes in the bone marrow and peripheral blood,48,49 we further explored the role of Atg5 within the eosinophil lineage in hypereosinophilic mice. Atg5eoΔII5tg mice were generated by crossing Atg5eoΔ mice with II5 (IL-5) transgenic mice overexpressing IL-5 in CD3+ T cells (NJ.1638).36 Knocking out Atg5 in the eosinophil lineage of Il5 transgenic mice also resulted in delayed and reduced eosinophil precursor proliferation and maturation (supplemental Figures 3-5). In addition, we found that in the absence of Atg5, Il5 transgenic mice exhibit reduced numbers of circulating eosinophils (supplemental Figure 4A) and reduced eosinophil tissue infiltration in liver, lungs, and partially also in spleen (supplemental Figure 4C-K). We also observed reduced expression of Gata-1, C/ebpε, Pu.1, and Trib1 in bone marrow–derived eosinophil precursors of Atg5eoΔII5tg mice (supplemental Figure 3H). Because this eosinophil population is heterogeneous regarding the differentiation status of individual cells, these alterations of transcription factor levels likely reflect the reduced and delayed eosinophil differentiation in the absence of Atg5.
Knocking out Atg5 in the eosinophil lineage results in reduced severity of FIP1L1-PDGFRα–mediated experimental eosinophilic leukemia
To further test the differentiation potential of Atg5-knockout eosinophils in mice under pathologic conditions, we used an established mouse model of chronic eosinophilic leukemia (CEL).50,51 A common clonal defect in CEL is caused by the constitutively activated tyrosine kinase FIP1L1-PDGFRα (F/P), a protein created by the fusion of the FIP1L1 and PDGFRα genes.52 In the experimental model used here, we purified hematopoietic stem cells/progenitors from the bone marrow of CtrlII5tg and Atg5eoΔII5tg donor mice that were transduced with the F/P fusion gene. Transduced cells were then transplanted into the wild-type recipient mice, in which the F/P–expressing cells engrafted and rapidly proliferated in the bone marrow.50,51 CEL has a latency of 4 to 5 weeks. We therefore euthanized CtrlII5tg-F/P and Atg5eoΔII5tg-F/P recipient mice on day 33 after transplantation and measured the blood counts. All recipients developed a strong leukocytosis, and CtrlII5tg-F/P recipients developed profound eosinophilia with increased B-cell counts (Figure 2A). Levels of eosinophils and B cells were decreased in Atg5eoΔII5tg-F/P recipient mice, whereas the number of neutrophils was strikingly elevated. The examination of blood smears revealed a high abundance of eosinophils in CtrlII5tg-F/P recipients, whereas neutrophils were the predominant leukocyte type in Atg5eoΔII5tg-F/P recipient mice (Figure 2B).
Knocking out Atg5 in the eosinophil lineage results in reduced severity of FIP1L1-PDGFRα–mediated experimental eosinophilic leukemia. (A) Cell counts. Total leukocyte counts were measured in the peripheral blood of CtrlII5tg-F/P and Atg5eoΔII5tg-F/P recipient mice using the scil Vet abc Plus+ hematology analyzer (scil animal care company GmbH, Viernheim, Germany) on day 33 after transplantation. Relative numbers of neutrophils (CD45+/CD11b+/Ly6G+), B cells (CD45+/B220+), T cells (CD45+/CD3+), monocytes (CD45+/CD11b+/CD115+), and eosinophils (CD45+/CD11b+/Siglec-F+) were determined by flow cytometry, and an absolute count for each cell population was calculated (n = 3-4). (B) Light microscopy. Peripheral blood smears of CtrlII5tg-F/P and Atg5eoΔII5tg-F/P recipient mice on day 33 after transplantation were stained with the Hemacolor Rapid Staining Kit (MilliporeSigma, Burlington, MA), followed by light microscopy analysis. Scale bars, 10 µm. (C) Flow cytometry. Relative numbers of cell populations transduced with the F/P fusion gene were determined by EGFP coexpression in the peripheral blood of CtrlII5tg-F/P and Atg5eoΔII5tg-F/P recipient mice 33 days after transplantation (n = 3-4). (D) Enzyme-linked immunosorbent assay measurements. IL-5 concentrations were measured in the plasma of CtrlII5tg-F/P and Atg5eoΔII5tg-F/P recipient mice on day 33 after transplantation (n = 3-4). Confocal microscopy. Formalin-fixed, paraffin-embedded liver (E) and colon (F) tissue sections of CtrlII5tg-F/P and Atg5eoΔII5tg-F/P recipient mice were stained with monoclonal mouse anti-EPX (red) and polyclonal goat anti-myeloperoxidase (MPO) (green) antibodies. Hoechst 33342 (blue) was used for nuclear DNA visualization. Quantification of EPX+ and MPO+ cells was performed by counting cells manually in 10 randomly selected high-power fields, each covering the area of 22.5 × 10−3 mm2 using an automatic digital slide scanner (Pannoramic MIDI II, 3DHistech, Budapest, Hungary) (n = 3). Right: representative confocal microscopy images are shown. Scale bars, 10 µm. Values are means ± standard error of the mean, or single data are presented in scatter dot plots in which the medians are indicated as red lines. *P < .05; **P < .01; ***P < .001. n.s., not significant.
Knocking out Atg5 in the eosinophil lineage results in reduced severity of FIP1L1-PDGFRα–mediated experimental eosinophilic leukemia. (A) Cell counts. Total leukocyte counts were measured in the peripheral blood of CtrlII5tg-F/P and Atg5eoΔII5tg-F/P recipient mice using the scil Vet abc Plus+ hematology analyzer (scil animal care company GmbH, Viernheim, Germany) on day 33 after transplantation. Relative numbers of neutrophils (CD45+/CD11b+/Ly6G+), B cells (CD45+/B220+), T cells (CD45+/CD3+), monocytes (CD45+/CD11b+/CD115+), and eosinophils (CD45+/CD11b+/Siglec-F+) were determined by flow cytometry, and an absolute count for each cell population was calculated (n = 3-4). (B) Light microscopy. Peripheral blood smears of CtrlII5tg-F/P and Atg5eoΔII5tg-F/P recipient mice on day 33 after transplantation were stained with the Hemacolor Rapid Staining Kit (MilliporeSigma, Burlington, MA), followed by light microscopy analysis. Scale bars, 10 µm. (C) Flow cytometry. Relative numbers of cell populations transduced with the F/P fusion gene were determined by EGFP coexpression in the peripheral blood of CtrlII5tg-F/P and Atg5eoΔII5tg-F/P recipient mice 33 days after transplantation (n = 3-4). (D) Enzyme-linked immunosorbent assay measurements. IL-5 concentrations were measured in the plasma of CtrlII5tg-F/P and Atg5eoΔII5tg-F/P recipient mice on day 33 after transplantation (n = 3-4). Confocal microscopy. Formalin-fixed, paraffin-embedded liver (E) and colon (F) tissue sections of CtrlII5tg-F/P and Atg5eoΔII5tg-F/P recipient mice were stained with monoclonal mouse anti-EPX (red) and polyclonal goat anti-myeloperoxidase (MPO) (green) antibodies. Hoechst 33342 (blue) was used for nuclear DNA visualization. Quantification of EPX+ and MPO+ cells was performed by counting cells manually in 10 randomly selected high-power fields, each covering the area of 22.5 × 10−3 mm2 using an automatic digital slide scanner (Pannoramic MIDI II, 3DHistech, Budapest, Hungary) (n = 3). Right: representative confocal microscopy images are shown. Scale bars, 10 µm. Values are means ± standard error of the mean, or single data are presented in scatter dot plots in which the medians are indicated as red lines. *P < .05; **P < .01; ***P < .001. n.s., not significant.
We further analyzed cell populations that were efficiently transduced with the F/P fusion gene, coexpressing the enhanced green fluorescent protein (EGFP). In CtrlII5tg-F/P recipient mice, eosinophils (CD45+/CD11b+/Siglec-F+) were the largest population among the circulating EGFP-expressing cells, confirming that F/P together with IL-5 overexpression promote eosinophil differentiation in vivo (Figure 2C). In contrast, Atg5eoΔII5tg-F/P recipients exhibited a striking decrease in EGFP-expressing eosinophils together with lower numbers of B cells, whereas the number of EGFP-expressing neutrophils was again significantly increased. In addition, there were no significant differences in T-cell engraftment between recipients.
Plasma IL-5 levels were high in both CtrlII5tg-F/P and Atg5eoΔII5tg-F/P recipient mice, but no differences were observed between the 2 genotypes, indicating that the differences in circulating eosinophil numbers were independent of IL-5 (Figure 2D). To show the effect of Atg5-knockout in eosinophils on granulocyte infiltration in the F/P mouse model of CEL, we stained liver and colon tissue sections with anti-EPX and antimyeloperoxidase antibodies. Decreased eosinophil and increased neutrophil infiltration was observed in the liver and colon of Atg5eoΔII5tg-F/P recipient mice (Figure 2E-F). These data are in agreement with our initial observations in peripheral blood (Figure 2A-C).
Collectively, our data suggest that Atg5eoΔII5tg-F/P recipient mice develop less severe eosinophilia, suggesting a restricted proliferation and differentiation of eosinophils in the absence of Atg5 also under the conditions of CEL.
Knocking out Atg5 in the eosinophil lineage results in increased bacterial clearance in Citrobacter rodentium–infected mice
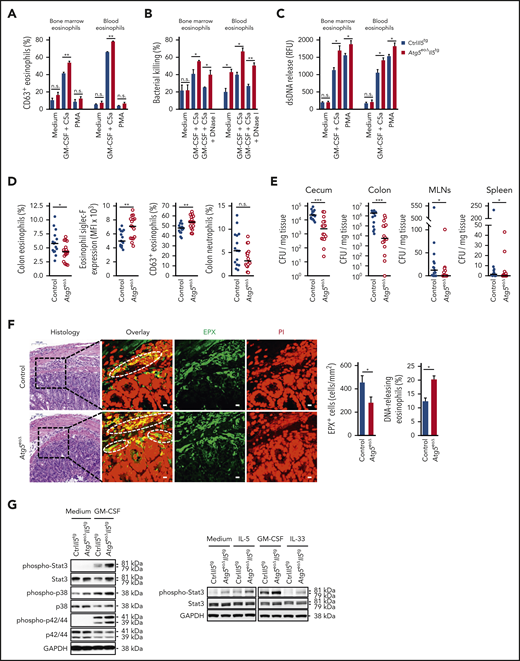
We hypothesized that the defect of autophagy in mature Atg5-knockout eosinophils causes defects in eosinophil effector functions. We therefore studied degranulation events in mouse eosinophils in the presence and absence of Atg5. Upon cell stimulation, intracellular CD63 is translocated to the cell periphery and plasma membrane; it is thus considered as a surrogate surface marker for eosinophil activation and degranulation.53,54 Atg5-knockout eosinophils, purified from the bone marrow or blood, revealed evidence of an enhanced degranulation, as we observed higher CD63 surface expression after GM-CSF priming and subsequent C5a stimulation compared with control eosinophils (Figure 3A). An increase in degranulation was observed with all C5a concentrations tested (supplemental Figure 6). In contrast to combined GM-CSF/C5a treatment, within the same time period, phorbol 12-myristate 13-acetate (PMA) had no effect on CD63 expression.
Knocking out Atg5 in the eosinophil lineage results in increased bacterial clearance in C rodentium–infected mice. (A) Flow cytometry. Eosinophils isolated from the bone marrow and peripheral blood of CtrlII5tg and Atg5eoΔII5tg mice were primed with recombinant mouse (rm) GM-CSF (25 ng/mL), followed by stimulation with rmC5a (10−8 M). Unprimed eosinophils were incubated with PMA (25 nM), and eosinophil degranulation was determined by measuring the increase in plasma membrane expression of surrogate marker CD63 (n = 4). (B) Flow cytometry. Freshly purified bone marrow and peripheral blood eosinophils of CtrlII5tg and Atg5eoΔII5tg mice were primed with rmGM-CSF, subsequently activated with rmC5a, and incubated with opsonized E coli–GFP in the presence or absence of DNase I. After the incubation, supernatants were collected, and the reduction of live bacteria was determined (n = 3). (C) Quantification of double-stranded DNA (dsDNA). Released dsDNA in the supernatants of isolated eosinophils from the bone marrow and peripheral blood of CtrlII5tg and Atg5eoΔII5tg mice after combined GM-CSF/C5a or PMA treatment was quantified by using PicoGreen fluorescent dye (Thermo Fisher Scientific, Waltham, MA) (n = 4). (D) Flow cytometry. Control and Atg5eoΔ mice were infected with C rodentium for 12 days, and relative numbers of infiltrated colonic eosinophils (CD45+/Siglec-F+) and neutrophils (CD45+/Ly6G+) were determined. Colonic lamina propria eosinophils from infected mice were analyzed for their activation and degranulation status by using markers Siglec-F and CD63, respectively (n = 15-16). (E) Bacterial clearance in vivo. Control and Atg5eoΔmice were infected with C rodentium, and the numbers of bacterial CFUs were assessed by plating homogenized cecum, colon, mesenteric lymph nodes (MLNs), and spleen on agar plates (n = 15-16). (F) Confocal microscopy. Colon tissues of control and Atg5eoΔ mice were obtained 12 days after infection with C rodentium. Hematoxylin and eosin staining images show the areas that were analyzed. Scale bars, 100 µm. Tissues were stained with monoclonal anti-EPX antibody (green) and propidium iodide (PI) (red). Representative confocal microscopy images are shown. White circles indicate colocalization of extracellular DNA and EPX. Scale bars, 10 µm. Right: quantification of EPX+ infiltrating eosinophils was performed by counting cells in 10 randomly selected high-power fields, each covering the area of 22.5 × 10−3 mm2 using the automatic digital slide scanner Pannoramic MIDI II. Quantification of the DNA-releasing eosinophils was determined manually (n = 5). (G) Immunoblotting. Left: bone marrow eosinophils of CtrlII5tg and Atg5eoΔII5tg mice were untreated or primed with rmGM-CSF for 5 minutes. Protein lysates were analyzed for phosphorylated Stat3 (Tyr705), phosphorylated p38 (Thr180/Tyr182), and phosphorylated p44/42 (Thr202/Tyr204) protein expression. Stat3, p38, p44/42, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) protein levels were analyzed as loading controls. Right: bone marrow eosinophils of CtrlII5tg and Atg5eoΔII5tg mice were untreated or primed with rmIL-5, rmGM-CSF, and rmIL-33 for 5 minutes. Protein lysates were analyzed for protein expression of phosphorylated Stat3 (Tyr705). Stat3 and GAPDH served as loading controls. Representative immunoblots of 2 independent experiments are shown. Values are means ± standard error of the mean, or single data are presented in scatter dot plots in which the medians are indicated as red lines. *P < .05; **P < .01; ***P < .001. n.s., not significant.
Knocking out Atg5 in the eosinophil lineage results in increased bacterial clearance in C rodentium–infected mice. (A) Flow cytometry. Eosinophils isolated from the bone marrow and peripheral blood of CtrlII5tg and Atg5eoΔII5tg mice were primed with recombinant mouse (rm) GM-CSF (25 ng/mL), followed by stimulation with rmC5a (10−8 M). Unprimed eosinophils were incubated with PMA (25 nM), and eosinophil degranulation was determined by measuring the increase in plasma membrane expression of surrogate marker CD63 (n = 4). (B) Flow cytometry. Freshly purified bone marrow and peripheral blood eosinophils of CtrlII5tg and Atg5eoΔII5tg mice were primed with rmGM-CSF, subsequently activated with rmC5a, and incubated with opsonized E coli–GFP in the presence or absence of DNase I. After the incubation, supernatants were collected, and the reduction of live bacteria was determined (n = 3). (C) Quantification of double-stranded DNA (dsDNA). Released dsDNA in the supernatants of isolated eosinophils from the bone marrow and peripheral blood of CtrlII5tg and Atg5eoΔII5tg mice after combined GM-CSF/C5a or PMA treatment was quantified by using PicoGreen fluorescent dye (Thermo Fisher Scientific, Waltham, MA) (n = 4). (D) Flow cytometry. Control and Atg5eoΔ mice were infected with C rodentium for 12 days, and relative numbers of infiltrated colonic eosinophils (CD45+/Siglec-F+) and neutrophils (CD45+/Ly6G+) were determined. Colonic lamina propria eosinophils from infected mice were analyzed for their activation and degranulation status by using markers Siglec-F and CD63, respectively (n = 15-16). (E) Bacterial clearance in vivo. Control and Atg5eoΔmice were infected with C rodentium, and the numbers of bacterial CFUs were assessed by plating homogenized cecum, colon, mesenteric lymph nodes (MLNs), and spleen on agar plates (n = 15-16). (F) Confocal microscopy. Colon tissues of control and Atg5eoΔ mice were obtained 12 days after infection with C rodentium. Hematoxylin and eosin staining images show the areas that were analyzed. Scale bars, 100 µm. Tissues were stained with monoclonal anti-EPX antibody (green) and propidium iodide (PI) (red). Representative confocal microscopy images are shown. White circles indicate colocalization of extracellular DNA and EPX. Scale bars, 10 µm. Right: quantification of EPX+ infiltrating eosinophils was performed by counting cells in 10 randomly selected high-power fields, each covering the area of 22.5 × 10−3 mm2 using the automatic digital slide scanner Pannoramic MIDI II. Quantification of the DNA-releasing eosinophils was determined manually (n = 5). (G) Immunoblotting. Left: bone marrow eosinophils of CtrlII5tg and Atg5eoΔII5tg mice were untreated or primed with rmGM-CSF for 5 minutes. Protein lysates were analyzed for phosphorylated Stat3 (Tyr705), phosphorylated p38 (Thr180/Tyr182), and phosphorylated p44/42 (Thr202/Tyr204) protein expression. Stat3, p38, p44/42, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) protein levels were analyzed as loading controls. Right: bone marrow eosinophils of CtrlII5tg and Atg5eoΔII5tg mice were untreated or primed with rmIL-5, rmGM-CSF, and rmIL-33 for 5 minutes. Protein lysates were analyzed for protein expression of phosphorylated Stat3 (Tyr705). Stat3 and GAPDH served as loading controls. Representative immunoblots of 2 independent experiments are shown. Values are means ± standard error of the mean, or single data are presented in scatter dot plots in which the medians are indicated as red lines. *P < .05; **P < .01; ***P < .001. n.s., not significant.
We then investigated the importance of Atg5 for the antibacterial activity of eosinophils in vitro. In agreement with increased degranulation capacity, eosinophils lacking Atg5 exhibited augmented bacterial killing of Escherichia coli-GFP compared with control eosinophils isolated from both bone marrow and blood (Figure 3B). These data support the surprising finding of increased degranulation of Atg5-knockout eosinophils and suggest that autophagy-defective eosinophils exhibit increased effector functions. Furthermore, the addition of DNase I lowered bacterial killing to the levels similar to those of unstimulated eosinophils in both genotypes, suggesting that the killing occurred, at least partially, with EETs.55 We previously reported that Atg5-knockout eosinophils are capable of forming EETs.35 A careful quantitative analysis revealed that Atg5-knockout eosinophils exhibit an even increased capacity of forming EETs compared with control eosinophils (Figure 3C), again pointing to an increased effector function of eosinophils in the absence of Atg5.
It has previously been shown that eosinophils are essential to clear a C rodentium infection of the gastrointestinal tract.56 We therefore used the C rodentium model to test the antibacterial defense of eosinophils under in vivo conditions. Control and Atg5eoΔ mice were infected orally with C rodentium for 12 days, and the infected mice were investigated for the frequencies of infiltrating colonic lamina propria eosinophils. Relative numbers of infiltrating eosinophils but not neutrophils were reduced in the colon of infected Atg5eoΔ mice (Figure 3D), similar to reduced eosinophil tissue infiltration in Atg5eoΔIl5tg mice. The infiltrating colonic eosinophils were further characterized for their activation and degranulation status by analyzing the surface expression of Siglec-F and CD63. Eosinophils of Atg5eoΔ mice exhibited higher activation and degranulation levels compared with control mice, supporting the in vitro observations that indicated increased degranulation of Atg5-knockout eosinophils (Figure 3A). Furthermore, we determined bacterial loads in cecum, colon, mesenteric lymph nodes, and spleen by counting plated bacterial colonies. Atg5eoΔ mice cleared an infection with C rodentium better than control mice in all organs, indicating that these mice are more capable of overcoming both local and systemic bacterial infections (Figure 3E).
To analyze the effect of Atg5 on the EET formation in response to bacterial infection in vivo, we investigated colon tissue sections after staining of DNA and eosinophil granules. As expected, infiltrating eosinophils in colon tissues of control mice formed EETs in response to C rodentium infection (Figure 3F). Despite decreased eosinophil infiltration in the infected colon, Atg5eoΔ mice exhibited an enhanced ability to form EETs (Figure 3F) and a reduced bacterial load in colon tissue under in vivo conditions (Figure 3E).
To understand the underlying mechanism of the enhanced effector function of Atg5-knockout eosinophils, we analyzed the signaling pathways of bone marrow–derived eosinophils from CtrlIl5tg and Atg5eoΔIl5tg mice after priming with GM-CSF. In the absence of Atg5, increased phosphorylation of Stat3, p38, and p44/42 (Figure 3G, left panel) was detected. In addition, increased Stat3 phosphorylation after IL-5 and IL-33 stimulation of Atg5-knockout eosinophils was observed, indicating that these cytokines also exert increased signaling responses (Figure 3G, right panel). Collectively, Atg5-knockout eosinophils exhibit evidence for a more efficient signal transduction after cytokine stimulation, contributing to increased degranulation, bacterial killing, and double-stranded DNA release in vitro.
ATG5 is required for the differentiation of human eosinophilic leukemic cells, and ATG5low-expressing human eosinophils are more susceptible to degranulation
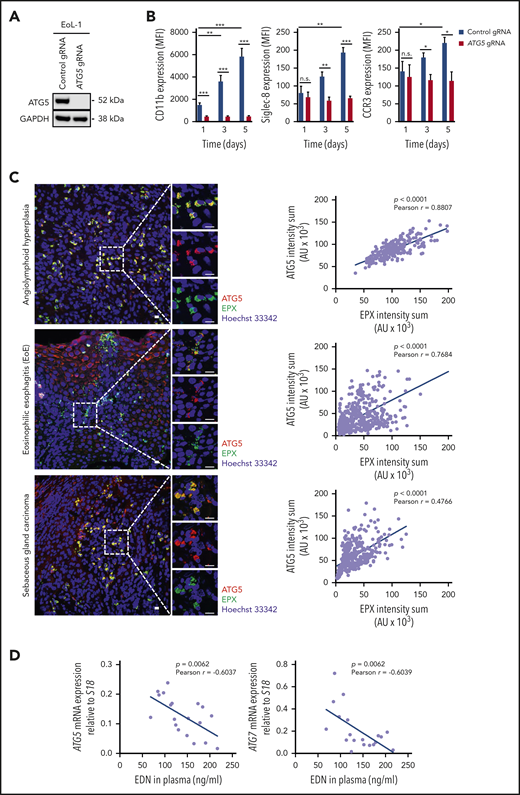
EoL-1 cells are derived from a patient with CEL and express FIP1L1-PDGFRα.57,58 We generated CRISPR/Cas9 guide RNA–mediated ATG5-knockout EoL-1 cells, which did not express ATG5 (Figure 4A). Upon induced differentiation of control and ATG5-knockout EoL-1 cells by 0.5 mM butyric acid,59 we observed a gradual increase in the surface expression of CD11b, Siglec-8, and CCR3 in control cells, whereas no increase of these markers was observed in ATG5-knockout EoL-1 cells (Figure 4B). Therefore, EoL-1 cells, as a model of an established human CEL, and F/P mice exhibited a decreased differentiation capacity of eosinophil precursors in the absence of ATG5.
ATG5 is required for the differentiation of human eosinophilic leukemic cells, and ATG5low-expressing human eosinophils are more susceptible to degranulation. (A) Immunoblotting. EoL-1 cells were transduced with lentiviral constructs containing Cas9 and guide RNA (gRNA) targeting ATG5. Whole-cell lysates of control and ATG5-knockout EoL-1 cells were collected and analyzed for the presence of ATG5 protein expression. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as loading control. Representative immunoblots of 2 independent experiments are shown. (B) Flow cytometry. Differentiation of control and ATG5-knockout EoL-1 cells was induced by the addition of 0.5 mM butyric acid, and the surface expression of eosinophil markers CD11b, Siglec-8, and CCR3 was assessed (n = 3). (C) Confocal microscopy. Formalin-fixed, paraffin-embedded human tissue sections of angiolymphoid hyperplasia (upper image), EoE (middle image), and sebaceous gland carcinoma (lower image) were stained with monoclonal mouse anti-EPX (green) and monoclonal rabbit anti-ATG5 (red) antibodies. Hoechst 33342 (blue) was used for nuclear DNA visualization. Representative confocal microscopy images are shown. Scale bars, 10 µm. Right: correlation analysis between the fluorescent intensity sums of EPX and ATG5, measured in all eosinophils, was performed for each tissue section, and the Pearson coefficient calculated in each case. (D) Correlation analysis between eosinophil-derived neurotoxin (EDN) levels in plasma and mRNA expression of ATG5 or ATG7 in blood eosinophils isolated from patients with hypereosinophilic syndrome (n = 19). Values are means ± standard error of the mean, or single data are presented in scatter dot plots in which the linear regression is indicated as blue lines. *P < .05; **P < .01; ***P < .001. AU, arbitrary unit; MFI, mean fluorescent intensity; n.s., not significant.
ATG5 is required for the differentiation of human eosinophilic leukemic cells, and ATG5low-expressing human eosinophils are more susceptible to degranulation. (A) Immunoblotting. EoL-1 cells were transduced with lentiviral constructs containing Cas9 and guide RNA (gRNA) targeting ATG5. Whole-cell lysates of control and ATG5-knockout EoL-1 cells were collected and analyzed for the presence of ATG5 protein expression. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as loading control. Representative immunoblots of 2 independent experiments are shown. (B) Flow cytometry. Differentiation of control and ATG5-knockout EoL-1 cells was induced by the addition of 0.5 mM butyric acid, and the surface expression of eosinophil markers CD11b, Siglec-8, and CCR3 was assessed (n = 3). (C) Confocal microscopy. Formalin-fixed, paraffin-embedded human tissue sections of angiolymphoid hyperplasia (upper image), EoE (middle image), and sebaceous gland carcinoma (lower image) were stained with monoclonal mouse anti-EPX (green) and monoclonal rabbit anti-ATG5 (red) antibodies. Hoechst 33342 (blue) was used for nuclear DNA visualization. Representative confocal microscopy images are shown. Scale bars, 10 µm. Right: correlation analysis between the fluorescent intensity sums of EPX and ATG5, measured in all eosinophils, was performed for each tissue section, and the Pearson coefficient calculated in each case. (D) Correlation analysis between eosinophil-derived neurotoxin (EDN) levels in plasma and mRNA expression of ATG5 or ATG7 in blood eosinophils isolated from patients with hypereosinophilic syndrome (n = 19). Values are means ± standard error of the mean, or single data are presented in scatter dot plots in which the linear regression is indicated as blue lines. *P < .05; **P < .01; ***P < .001. AU, arbitrary unit; MFI, mean fluorescent intensity; n.s., not significant.
We next analyzed ATG5 expression in human eosinophils and their degranulation in human eosinophilic tissues. Tissue eosinophils from a patient with angiolymphoid hyperplasia exhibited a strong correlation between ATG5 expression and intracellular EPX levels (Figure 4C, upper panels). Previously published work suggested a high degree of eosinophil degranulation and extracellular EPX deposition in EoE.60 In agreement with these findings, intracellular EPX levels in tissue eosinophils of a patient with EoE were relatively low. Strikingly, ATG5 expression was also low in the majority of the infiltrating eosinophils. Moreover, despite having a large population of ATG5low-expressing and degranulating eosinophils, there was a considerable heterogeneity among eosinophils that again showed a positive correlation between ATG5 expression and intracellular EPX levels (Figure 4C, middle panels). A similar situation was seen in a third patient with a sebaceous gland carcinoma with associated tissue eosinophilia in which we also observed a large population of ATG5low-expressing and degranulating eosinophils; this finding was similar to that seen in the patient with EoE (Figure 4C, lower panels).
We next examined the mRNA expression of ATG5 and ATG7 in blood eosinophils from 19 patients with hypereosinophilic syndrome and measured the concentration of eosinophil-derived neurotoxin in the plasma of the same patients. A significant negative correlation was observed between the expression of ATG5 (and ATG7) in blood eosinophils and eosinophil-derived neurotoxin levels in plasma (Figure 4D). These data support the concept that ATG5low-expressing eosinophils are prone to enhanced degranulation.
Discussion
Eosinophils have historically been described as major effector cells in the host defense against helminth infections, causing tissue damage owing to the release of their highly toxic granule proteins. Recent data suggest that eosinophils are not only involved in immunologic effector functions but also perform tissue-protective and immunoregulatory functions. For instance, eosinophils are now recognized for their control of innate and adaptive immune responses.1 Despite the clinical significance of eosinophils, little information is available regarding mechanisms underlying eosinophil hematopoiesis. There is an urgent need to dissect the molecular mechanism of eosinophil generation, partly because of their pathogenic role in allergic and autoimmune disorders,61 but also because of the importance of eosinophils in immune regulation and antibacterial defense mechanisms.1
A role for autophagy has been suggested for neutrophil differentiation,21,22 whereas no research was conducted to examine involvement of autophagy in eosinophil differentiation. To investigate the potential role of autophagy within the eosinophil lineage, we used a genetic approach and created mice with an eosinophil-specific knockout of Atg5. We newly report a decreased proliferation and delayed maturation of in vitro differentiating Atg5-knockout eosinophils. Atg5eoΔ and Atg5eoΔII5tg mice had an enlarged pool of immature eosinophils compared with control mice. We detected no cell death abnormalities in Atg5-knockout eosinophils, excluding a role for increased eosinophil death as reported for the proapoptotic activities of paired immunoglobulin-like receptor A.15 As a consequence of reduced eosinophil differentiation, we measured substantially reduced counts of mature eosinophils in the blood and peripheral tissues of Atg5eoΔII5tg mice compared with CtrlII5tg mice. ATGs have been shown to regulate transcription factors.62 In the absence of Atg5, we observed reduced expression of the transcription factors Gata-1, C/ebpε, Pu.1, and Trib1, although these changes may be an effect of delayed and reduced eosinophil hematopoiesis rather than the cause of impaired differentiation.
It should be noted that the effects of Atg5-knockout in neutrophil differentiation are not in agreement with the findings reported here. The absence of Atg5 in the neutrophil lineage resulted in increased proliferation of neutrophil precursor cells and an accelerated process of neutrophil differentiation, resulting in an accumulation of neutrophils in bone marrow, blood, spleen, and lymph nodes.21 Although the reason for these surprising differences between eosinophil and neutrophil differentiation remains to be investigated, a single-cell transcriptome analysis identified an early hematopoietic-lineage bifurcation that separates the myeloid lineages.63 One population, expressing GATA-1, can be differentiated into eosinophils, mast cells, megakaryocytes, and erythroid cells. The other GATA-1–negative population can generate neutrophils, monocytes, and lymphocytes. It is possible that autophagy in these 2 myeloid differentiation pathways is differentially regulated. It has also been reported that TRIB1 expression promotes eosinophil identity by restraining neutrophil lineage commitment, partly by decreasing levels of C/EBPα.64 We observed reduced Trib1 and increased C/ebpα expression in eosinophil precursors from Atg5eoΔII5tg mice, a finding that might partially explain the increased neutrophil population in Atg5eoΔII5tg-F/P recipient mice.
We also observed a decline in circulating B cells and monocytes in these mice. Our observations are in agreement with an earlier study in which eosinophils were shown to promote B-cell survival, proliferation, and immunoglobulin secretion.65 It was also reported that eosinophils support the survival of plasma cells by secreting the proliferation-inducing ligands APRIL and IL-6.66,67 A recent study, however, argued against a role of eosinophils in the maintenance of the plasma cells and challenged the concept of an eosinophil‐sustained bone marrow niche.68 It would therefore be interesting to measure the secretion of immunoglobulin G from bone marrow cells, including the expression of the plasma cell survival factors, APRIL and IL-6, in eosinophils of Atg5eoΔII5tg compared with CtrlII5tg mice. The decline in monocytes in Atg5eoΔIl5tg mice might be the consequence of decreased eosinophil and/or B-cell numbers that can produce monocyte survival factors such as IL-6 and M-CSF.69,70
Degranulation represents one of the key effector functions of eosinophils, which preserve their ability to respond to consecutive stimuli, even after repeated activation with the same ligand.71 Autophagy was shown to play a crucial role in the degranulation of murine mast cells72 and neutrophils.73 Mice with an autophagy deficiency in the myeloid cell lineage exhibited reduced severity of several neutrophil-mediated inflammatory and autoimmune disease models, including PMA-induced ear inflammation, lipopolysaccharide-induced breakdown of the blood–brain barrier, and experimental autoimmune encephalomyelitis. The most likely mechanism was the reduced NADPH oxidase-mediated production of reactive oxygen species in Atg7-knockout neutrophils.73 Intriguingly, our findings show an increased capacity of Atg5-knockout eosinophils to degranulate and form functional EETs, suggesting contrasting effects of autophagy deficiency not only on differentiation but also on effector functions in neutrophils and eosinophils. Previously published work has shown the importance of p38 MAPK and extracellular signal–regulated kinase activities for eosinophil degranulation.43 We observed increased phosphorylation of Stat3, p38, and p44/42 in Atg5-knockout eosinophils after GM-CSF priming. Conversely, both p38 and p44/42 activities were reportedly reduced during the terminal differentiation process in the absence of Atg5. We speculate that MAPK activities are differentially regulated upon full maturation, resulting in an increased susceptibility to GM-CSF stimulation, which could explain the enhanced effector functions. Because eosinophils are a common tumor infiltrate and can limit tumor growth through the release of toxic granule proteins,74 it would be interesting to investigate tumoricidal activity of Atg5-knockout eosinophils.
We also obtained evidence for a role of ATG5 in eosinophil hematopoiesis and degranulation in human in vitro and in vivo systems, suggesting a conserved function of ATG5 in mouse and human eosinophils. Taken together, our findings indicate an involvement of Atg5-dependent autophagy in the process of eosinophil hematopoiesis, whereby autophagy seems to positively regulate proliferation and differentiation of eosinophil precursors. Consequently, Atg5-knockout eosinophils develop less severe CEL in vivo. Moreover, eosinophils lacking Atg5 exhibit increased degranulation and better bacterial clearance, indicating that autophagy negatively regulates eosinophil effector functions. It should be noted, however, that ATG5 can also mediate autophagy-independent functions.75 Therefore, despite our demonstration that Atg5-knockout eosinophils are autophagy deficient, some of the observed effects described here might be ATG5 dependent but autophagy independent. Nevertheless, targeting ATG5 in eosinophils might be a useful therapeutic strategy in eosinophil-associated cancers, including eosinophilic leukemias, and bacterial infections in which eosinophils are important effector cells.
Requests for data may be sent to the corresponding author at hus@pki.unibe.ch.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are indebted to the many clinicians who permitted analysis of their eosinophilic patients. The authors thank C. Münz (University of Zürich, Zurich, Switzerland) for kindly providing Atg5flox/flox mice and J. J. Lee (Mayo Clinic, Phoenix, AZ) for eoCre and Il5tg mice. They also thank Evelyne Kozlowski for technical support with the confocal microscopy experiments.
This work was supported by the Swiss National Science Foundation (grant 310030_184816 [H.-U.S.]; 31003A_173215 [S.Y.]). The authors also acknowledge financial support by the Russian Government Program “Recruitment of the Leading Scientists into the Russian Institutions of Higher Education” (H.-U.S.). N.G. and A.H. are students of the Graduate School of Cellular and Biomedical Sciences of the University of Bern. Images were acquired on equipment supported by the Microscopy Imaging Centre of the University of Bern.
Authorship
Contribution: N.G. conceived, planned and performed the study, analyzed and interpreted data, and wrote the paper; A.H., D.S., M.C., and S.C. performed experiments; K.O., C.B., I.C.A., and C.R. supported the in vivo mouse experiments; A.A.-S. and A.M. helped with the design of the in vivo mouse experiments and edited the paper; S.Y. provided experimental advice, performed experiments, and edited the paper; H.-U.S. provided overall guidance, experimental advice, and the laboratory infrastructure, and also edited the paper; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hans-Uwe Simon, Institute of Pharmacology, University of Bern, Inselspital, INO-F, CH-3010 Bern, Switzerland; e-mail: hus@pki.unibe.ch.

![Knocking out Atg5 in the eosinophil lineage results in delayed and reduced eosinophil precursor proliferation and maturation. (A) Flow cytometry. Maturity of eosinophils in the bone marrow of control and Atg5eoΔ mice was assessed by measuring the cell surface expression of chemokine receptor CCR3. Mature eosinophils were defined as Siglec-F+CCR3+ and immature eosinophils as Siglec-F+CCR3– cell populations (n = 6). Right: representative original flow cytometry data are shown. Each symbol represents a value for an individual mouse throughout. (B) The data of panel A are presented as absolute numbers. (C) Flow cytometry. Eosinophils were differentiated in vitro from bone marrow cells of control and Atg5eoΔ mice, and their maturity was assessed by measuring the surface expression of CCR3 on the indicated days (n = 3). Right: representative original flow cytometry data are shown. (D) Flow cytometry. Proliferating eosinophil precursors in the S phase of the cell cycle during in vitro differentiation were detected by 5-ethynyl-2'-deoxyuridine (EdU) labeling (n = 3). (E) Flow cytometry. The proliferative status of eosinophil precursors in the G1, S, G2, and M phases of the cell cycle during in vitro differentiation was determined by Ki-67 expression measurements (n = 3). (F) [3H]‐thymidine incorporation assay. For each time point, [3H]‐thymidine was added to the cultures of in vitro differentiating eosinophils 24 hours before the measurement. Radioactivity in DNA recovered from the cells was determined with a scintillation counter to determine the extent of cell division (n = 3). (G) CFU assay. Freshly isolated bone marrow cells from control and Atg5eoΔ mice were plated in methylcellulose media supplemented with recombinant mouse IL-5. After 12 days of incubation, cells were stained with the substrate for EPX, and brown-stained Eo-CFUs of >50 cells were counted in each well (n = 5). Right: representative images of culture wells and single colonies are shown. Scale bars, 10 µm. To confirm the identification of colonies as Eo-CFU, samples were selected and removed from the wells for analysis of Siglec-F expression by flow cytometry as presented in supplemental Figure 1H. (H) Immunoblotting. Protein lysates during in vitro culture of differentiating eosinophils were collected on the indicated days and analyzed for the presence of phosphorylated p38 (Thr180/Tyr182), phosphorylated p44/42 (Thr202/Tyr204), ATG5, and LC3 protein expression. p38, p44/42, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) protein levels were analyzed as loading controls. Representative immunoblots of 3 independent experiments are shown. Values are means ± standard error of the mean, or single data are presented in scatter dot plots in which the medians are indicated as red lines. *P < .05; **P < .01; ***P < .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/137/21/10.1182_blood.2020010208/1/m_bloodbld2020010208f1.png?Expires=1769628206&Signature=ajk~NKBSGLvwjN0NNHe~sgsVtoz6AMrWH29IdKlur0CzgrPoOYxdtl-6o0isYjeo43fvEbRi9kWRh1SEtctZAr5zBiSHHEPdHHUyD6QbZbUPQnPy-WZThfBdFuFADqQzpHbUmxrErr~BicwyuOHfa3~u3iDcOX1SPBYSEmzXEiBofVXgkK-TBmCQ5WvSe3WhNTuSwDVfbdoE91dGEZo7hh~YxXpnFv~Dp45iol0KGNv6O5L3y7uStCET31Nm7z5blCfmOcUCgnzBCqpDj1IfyW-Mc5tAefbmzkuC~eRQxAvlXTvFDDUzvIsm7oUJDQCtXEABHDvDVMyLsS0llsF~Gw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal