RESPONSE
We thank Abdo and colleagues1 for their interest in our recent publication2 and for their insights into the cell of origin (COO) of TCF3-ZNF384 and PTPN11 mutations in monozygotic twins with concordant B-cell progenitor-acute lymphoblastric leukemia (BCP-ALL). We are delighted that the conclusions reached by their independent analyses are suggestive of the same underlying conclusions that we reached: the inferred COO is around the onset of immunoglobulin heavy chain (IgH) DJ rearrangement in the bone marrow, likely in a pre-DJ progenitor. Abdo et al raise some important points about the study upon which we can comment and elaborate (Figure 1A).
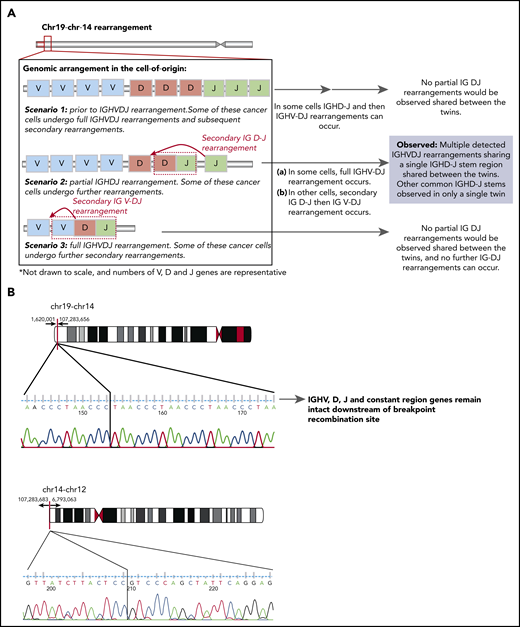
Schematic diagram of the possible IGH locus rearrangements present in the (COO) and the genomic breakpoints of the shared t(14;19) and t(12;14). (A) Scenario of the possible IGH locus rearrangements present in the (COO). Scenario 1 has the COO at the stage prior to IGH VDJ rearrangement. Some of these cancer cells may undergo full IGH VDJ rearrangements and subsequent secondary rearrangements, however these would not be shared between the twins. Scenario 2 has the COO at the stage with a partial IGH DJ rearrangement that is shared between the twins. Some of these cancer cells can undergo further IG V- DJ and/or secondary rearrangements (where a primary immunoglobulin DJ rearrangement can be replaced by an upstream D to downstream J) that may be present only in 1 twin. Scenario 3 has the COO with a full IgH VDJ rearrangement. Some of these cancer cells may undergo further secondary immunoglobulin V rearrangements. Only Scenario 2 is supported by the data. (B) Schematic representation of the genomic breakpoints determined by whole-genome sequencing at the base pair level for t(14;19) and t(12;14).
Schematic diagram of the possible IGH locus rearrangements present in the (COO) and the genomic breakpoints of the shared t(14;19) and t(12;14). (A) Scenario of the possible IGH locus rearrangements present in the (COO). Scenario 1 has the COO at the stage prior to IGH VDJ rearrangement. Some of these cancer cells may undergo full IGH VDJ rearrangements and subsequent secondary rearrangements, however these would not be shared between the twins. Scenario 2 has the COO at the stage with a partial IGH DJ rearrangement that is shared between the twins. Some of these cancer cells can undergo further IG V- DJ and/or secondary rearrangements (where a primary immunoglobulin DJ rearrangement can be replaced by an upstream D to downstream J) that may be present only in 1 twin. Scenario 3 has the COO with a full IgH VDJ rearrangement. Some of these cancer cells may undergo further secondary immunoglobulin V rearrangements. Only Scenario 2 is supported by the data. (B) Schematic representation of the genomic breakpoints determined by whole-genome sequencing at the base pair level for t(14;19) and t(12;14).
First, the complex rearrangement leading to the TCF3-ZNF384 fusion resulted from a 3-way t(12;14) and t(14;19) translocation, and we confirmed that the chromosome 14 breakpoint involved the end of the IgH locus, spanning from position 107 283 ,683 onward (the whole locus encompasses position 106,03 2614 to position 107 288,051). Chromosomal breakpoints are identical in the twins, as depicted in the graphical representation of the breakpoint (Figure 1B). Involvement of the IgH locus in rearrangements is well recognized in BCP-ALL.3
Second, quantification of IgH VDJ rearrangements by Abdo et al demonstrated 2 or 3 significantly clonal IgH VDJ rearrangements (comprising >10% of B-cell receptors [BCRs]) within each twin, some of which share a common DJ incomplete rearrangement. This observation was not observed in our BCR data for 2 reasons: first, we performed BCR sequencing analysis on RNA, whereas Abdo and colleagues performed their analyses on DNA; analyses can provide different results that are dependent on RNA expression of leukemic cells and the presence of nonexpressed allele defective rearrangements present in the genomic DNA.3 A second, and more significant, difference between the 2 approaches was that out-of-frame nonfunctional BCRs were filtered out from our dataset to reduce the potential of retaining reads with sequencing or polymerase chain reaction errors, which is an accepted practice for analyzing functional BCRs.4 As pointed out by Abdo and colleagues, the common rearrangements from these twins were out of frame.
Therefore, a reanalysis of our data with the modified filtering criteria, including all BCRs, and irrespective of the presence of stop codons, was performed. In accordance with Abdo and colleagues, 57.24% and 54% of the BCRs from this new filtering strategy contained stop codons for Twin A and Twin B, respectively (Table 1). From this, we could identify 3 expanded clones from Twin A matching the descriptions from Abdo et al and 2 clones from Twin B (Table 2). These IgH VDJ rearrangements are all out of frame, containing the stop codon within the conserved imputed IGHD region of the rearrangements.
Total numbers of BCRs after different filtering strategies
| Sample . | Excluding BCRs with stop codons, n . | Including BCRs with stop codons, n . | BCRs with stop codons, % . |
|---|---|---|---|
| Twin A | 9 562 | 22 362 | 57.24 |
| Twin B | 22 790 | 49 541 | 54.00 |
| Sample . | Excluding BCRs with stop codons, n . | Including BCRs with stop codons, n . | BCRs with stop codons, % . |
|---|---|---|---|
| Twin A | 9 562 | 22 362 | 57.24 |
| Twin B | 22 790 | 49 541 | 54.00 |
Characterization of clones representing more than 2% in the BCR repertoires (including stop codons)
| . | Percentage of repertoire . | V gene . | D gene . | J gene . | SHM in V gene . | STOP codon present? . | CDR3 sequence (nucleotides) . | Predicted CDR3 sequence based on in-frame IGHV gene (amino acids) . |
|---|---|---|---|---|---|---|---|---|
| Twin A | 14.95 | IGHV3-33*01/06 | IGHD7-27*01 | IGHJ4*02 | 0 | Yes | GCGATCCTAACTGGGGACCTAGTACTACTTTGACTAC | AILTGDLVLL*L |
| Twin A | 14.41 | IGHV3- 9*01 | IGHD6-13*01 | IGHJ4*02 | 0 | Yes | GCAAAAGATATAAGGGGTAGCAGCAGCTTTTGACTAC | AKDIRGSSSF*L |
| Twin A | 2.45 | IGHV3-21*01/02 | IGHD6-13*01 | IGHJ4*02 | 0 | Yes | GCGAGAGATCCGCCGTAGCAGCAGCTTTTGACTAC | ARDPP*QQLLTT |
| Twin B | 28.36 | IGHV3-74*01/02 | IGHD6-13*01 | IGHJ4*02 | 0 | Yes | GCAACCCGTGGGAGCAGCAGCTTTTGACTAC | ATRGSSSF*L |
| Twin B | 3.12 | IGHV3-30*04/03 | IGHD6-13*01 | IGHJ4*02 | 0 | Yes | GCGAGAGATCCCCCCTCCGTATAGCAGCAGCTTTTGACTAC | ARDPPSV*QQLLTT |
| . | Percentage of repertoire . | V gene . | D gene . | J gene . | SHM in V gene . | STOP codon present? . | CDR3 sequence (nucleotides) . | Predicted CDR3 sequence based on in-frame IGHV gene (amino acids) . |
|---|---|---|---|---|---|---|---|---|
| Twin A | 14.95 | IGHV3-33*01/06 | IGHD7-27*01 | IGHJ4*02 | 0 | Yes | GCGATCCTAACTGGGGACCTAGTACTACTTTGACTAC | AILTGDLVLL*L |
| Twin A | 14.41 | IGHV3- 9*01 | IGHD6-13*01 | IGHJ4*02 | 0 | Yes | GCAAAAGATATAAGGGGTAGCAGCAGCTTTTGACTAC | AKDIRGSSSF*L |
| Twin A | 2.45 | IGHV3-21*01/02 | IGHD6-13*01 | IGHJ4*02 | 0 | Yes | GCGAGAGATCCGCCGTAGCAGCAGCTTTTGACTAC | ARDPP*QQLLTT |
| Twin B | 28.36 | IGHV3-74*01/02 | IGHD6-13*01 | IGHJ4*02 | 0 | Yes | GCAACCCGTGGGAGCAGCAGCTTTTGACTAC | ATRGSSSF*L |
| Twin B | 3.12 | IGHV3-30*04/03 | IGHD6-13*01 | IGHJ4*02 | 0 | Yes | GCGAGAGATCCCCCCTCCGTATAGCAGCAGCTTTTGACTAC | ARDPPSV*QQLLTT |
SHM, somatic hypermutation.
In agreement with Abdo et al, we identify a major DJ rearrangement that is common to the most frequent clones in both twins; it consists of IGHD6-13*01 and IGHJ4*02 but different IGHV genes (Table 2; supplemental Figure 1, available on the Blood Web site). This rearrangement does not contain any N nucleotide additions. Indeed, the presence of a common DJ and multiple IGHV genes is suggestive of secondary rearrangements, which are very common in B-ALL.5,6 Secondary rearrangements are thought to be a result of aberrant activity of recombination-activating genes promoting genomic rearrangements critical to B-cell acute lymphoblastic leukemia (ALL) pathogenesis.7,8 IgHD-J combinations (including junctional regions), known as “stem sequences,” are stable in instances of secondary rearrangements.9 The false-positive detection rates3 for these secondary rearrangements is estimated to be 9.245 × 10−6.
The largest clone in Twin A contains the IGHD7-27*01 and IGHJ4*02 rearrangement (14.95% of BCRs and also including a stop codon) and contains N nucleotide additions; it was also observed by Abdo et al at a high frequency. Here, we also detected multiple clones related by secondary rearrangement sharing the stem DJ sequence in Twin A (supplemental Figure 2). It appears that these 2 expanded clones are, in fact, from distinct rearrangements rather than being derived from a common pre-DJ precursor. Indeed, IGHJ4 is the most commonly rearranged IGHJ gene (comprising ∼50% of BCRs from healthy individuals10 ); therefore, the common IGHJ rearrangement may have occurred independently by chance.
Abdo et al suggest that the polyclonal BCR repertoires that we described are likely to correspond to nonleukemic IgH VDJ rearrangements. However, BCR repertoires were assessed for highly purified (purity > 99%) CD34+CD19+CD10− pro-B acute lymphoblastic leukemia (ALL) cells from peripheral blood. Such pro-B normal counterparts do not exist in peripheral blood, ruling out the possibility that polyclonal BCR repertories are nonleukemic. The aforementioned presence of frequent and ongoing secondary rearrangements in cells containing both of these DJ rearrangements provides further evidence that these are both derived from bona fide ALL cells and that there are multiple DJ rearrangements associated with the leukemic cell population. Given that there is only a single IgH DJ stem region shared by both twins, this suggests that the COO must be prior to the full IgH VDJ rearrangement and after the IgH DJ rearrangement. In support of this, the expanded ALL clones observed in only 1 twin containing distinct IgH DJ stem regions may arise from a primary IG DJ rearrangement that can be replaced by secondary recombination of an upstream D to downstream J.11 This means that a polyclonal repertoire of distinct IgH VDJ rearrangements may be generated from an ALL population containing a partial immunoglobulin DJ rearrangement (Figure 1A). Data from Abdo et al support our previous conclusion that the predicted COO shared by the 2 twins is at the IgH DJ recombination stage, rather than the containing an unrecombined (germline) or fully recombined IGH locus, as the COO of TCF3-ZNF384 and PTPN11 mutations.
Data sharing requests should be sent to Pablo Menendez (pmenendez@carrerasresearch.org).
The online version of this article contains a data supplement.
Authorship
Contribution: R.B.-R. analyzed data and C.B., P.B., I.V., P.M. and R.B.-R. contributed to the artwork and wrote the manuscript.
Conflict-of-interest disclosure: R.B.-R. is a cofounder of and consultant for Alchemab Therapeutics Ltd and has been a consultant for Imperial College London and VHSquared. The remaining authors declare no competing financial interests.
Correspondence: Pablo Menendez, Josep Carraras Leukemia Research Institute, Carrer Casanova 143, Facultat de Medicina, University of Barcelona, Barcelona 08036, Spain; e-mail: pmenendez@carrerasresearch.org; and Rachael Bashford-Rogers, Wellcome Centre for Human Genetics, University of Oxford, Oxford, United Kingdom; e-mail: rbr1@well.ox.ac.uk.