Key Points
Many properties are shared between the lamprey and human NPRL3-linked HB locus, including a remote erythroid enhancer in intron 7 of NPRL3.
Linkage of multiple globin genes to the same adjacent gene explains how hemoglobins could undergo convergent evolution in different species
Abstract
The oxygen transport function of hemoglobin (HB) is thought to have arisen ∼500 million years ago, roughly coinciding with the divergence between jawless (Agnatha) and jawed (Gnathostomata) vertebrates. Intriguingly, extant HBs of jawless and jawed vertebrates were shown to have evolved twice, and independently, from different ancestral globin proteins. This raises the question of whether erythroid-specific expression of HB also evolved twice independently. In all jawed vertebrates studied to date, one of the HB gene clusters is linked to the widely expressed NPRL3 gene. Here we show that the nprl3-linked hb locus of a jawless vertebrate, the river lamprey (Lampetra fluviatilis), shares a range of structural and functional properties with the equivalent jawed vertebrate HB locus. Functional analysis demonstrates that an erythroid-specific enhancer is located in intron 7 of lamprey nprl3, which corresponds to the NPRL3 intron 7 MCS-R1 enhancer of jawed vertebrates. Collectively, our findings signify the presence of an nprl3-linked multiglobin gene locus, which contains a remote enhancer that drives globin expression in erythroid cells, before the divergence of jawless and jawed vertebrates. Different globin genes from this ancestral cluster evolved in the current NPRL3-linked HB genes in jawless and jawed vertebrates. This provides an explanation of the enigma of how, in different species, globin genes linked to the same adjacent gene could undergo convergent evolution.
Introduction
Hemoglobin (HB) is responsible for the oxygen transport function of erythrocytes and comprises more than 90% of the soluble protein in these cells. Each erythrocyte contains approximately 250 × 106 HB molecules. To attain these high numbers, expression of the HB genes is activated by powerful distal erythroid-specific enhancers.1 Given the importance of HB in human physiology and its role in hemoglobinopathies such as α-thalassemia, β-thalassemia, and sickle cell disease, the structure and evolutionary origin of HB loci and proteins have been intensively studied.2,3 Globin-related proteins are present in all phyla of life.4 Because the first single-celled organisms were anaerobic, the original function of globins was most likely in detoxification, acting as oxygen scavengers and peroxidases or deoxygenases.5,6 As aerobic multicellular organisms evolved and increased in size, they became dependent on globins to provide an oxygen transport and storage system. In extant mammals, the monomeric myoglobin (MB) is still used as an oxygen storage protein in muscle.7 The oxygen transport function of HB is thought to have arisen ∼500 million years ago,8 roughly coinciding with the divergence between jawless (Agnatha) and jawed (Gnathostomata) vertebrates.9 Cyclostomata (lampreys and hagfish) are extant representatives of jawless vertebrates, and therefore characterization of these species may provide important insights into the evolutionary origins of vertebrate genomes, loci, and proteins.10 It is widely accepted that there is a common origin of HBs from a proto-HB protein that evolved to become an oxygen transporter in the common ancestor of all vertebrates.4,11 However, by contrast, recent phylogenetic analyses concluded that HBs arose twice, and independently, from different ancestral globin proteins in jawless and jawed vertebrates.8,12,13 This raises the question of whether erythroid-specific expression of the HB genes also evolved twice. In jawed vertebrates, one of the HB gene clusters is linked to the widely expressed NPRL3 gene. When investigated, distal erythroid regulatory elements that activate the linked HB genes are invariably present in introns of the NPRL3 gene.14,15 The two strongest enhancers, called regulatory multispecies conserved sequences or MCS-Rs, are MCS-R1 and MCS-R2.16 To further investigate the evolutionary origin of vertebrate HB loci, we isolated and sequenced 2 cosmids covering the hb cluster linked to the nprl3 gene in the river lamprey (Lampetra fluviatilis), a jawless vertebrate. Our analysis demonstrates that this L fluviatilis hb locus shares an uncanny range of structural and functional properties with the jawed vertebrate NPRL3-linked HB locus. Chromatin accessibility mapping and functional analysis demonstrate that an erythroid-specific enhancer is located in intron 7 of lamprey nprl3, which corresponds to the NPRL3 intron 7 MCS-R1 enhancer of jawed vertebrates. We infer that multiple globin genes may have been linked to NPRL3 before the divergence of jawless and jawed vertebrates, explaining how, in different species, globin genes linked to the same adjacent gene could undergo convergent evolution.
Materials and methods
Fish
Adult European river lampreys (L fluviatilis [taxonomy ID, 7748]) were freshly caught from the Kymijoki river in southeast Finland. Tissues were collected and immediately frozen. Lamprey larvae (ammocetes) were preserved in 70% ethanol.
Migratory-phase adult sea lampreys (Petromyzon marinus [taxonomy ID, 7757]) were trapped in rivers in Michigan by the Great Lakes Fisheries Commission and shipped overnight from United States Geological Survey Hammond Bay Biological Station. Sea lamprey ammocetes were captured and shipped by Lamprey Services (Ludington, MI). All sea lamprey were imported in accordance with a detrimental species permit approved by the California Department of Fish and Wildlife and were maintained in tanks in accordance with the Caltech Institutional Animal Care and Use Committee protocol #1436.
L fluviatilis cosmid library
Arrayed filters of an L fluviatilis cosmid library were obtained from the German Science Centre for Genome Research (Berlin, Germany). Isolation and characterization of cosmids containing hb genes are described in supplemental Materials and methods (available on the Blood Web site). Two partially overlapping cosmids (99E08 and 109N16) were selected for sequence analysis.
Sequencing and contig assembly
Cosmid DNA was sheared into ∼2-kb fragments, which were used for shotgun cloning and Sanger sequencing. Paired-end sequences were used to construct contigs (see supplemental Materials and methods). Potential exons and genes were identified by GenScan,17 and these initial gene structures were further refined by manual curation. Predicted protein sequences were used to search the genome databases at National Center for Biotechnology Information (NCBI) and Ensembl with the Basic Local Alignment Search Tool (BLAST) family of search algorithms.18,19
Long-range amplification of genomic DNA and Nanopore sequencing
Long-range polymerase chain reaction (PCR) using genomic L fluviatilis genomic DNA as a template was used to generate 5 overlapping amplicons. Primers and annealing temperatures are listed in supplemental Table 2. Amplifications were performed using Prime STAR GXL DNA polymerase (TaKaRa Bio Inc, Shiga, Japan). Between 1.0 and 1.5 μg of amplicons were pooled to sequence with the Nanopore 1D Amplicon Sequencing strategy for the MinION using kit SQK-LSK108 (Oxford Nanopore Technologies, Oxford, United Kingdom). MinION sequencing was performed according to the manufacturer’s guidelines using an R9 flow cell (FLO-MIN106). Additional details regarding long-range amplification and alignment and contig assembly are described in supplemental Materials and methods.
Comparative analysis
Assemblies of the L fluviatilis hb locus based on Sanger and Nanopore sequencing data were aligned and visualized using PipMaker20 with default settings. Globin and NPRL3 protein sequences were retrieved from the NCBI and Ensembl databases. Protein sequences are listed in supplemental Information. Multiple sequence alignments and molecular phylogenetic analyses are described in supplemental Materials and methods. Genomic sequences of the pufferfish and human NPRL3 genes were retrieved from Ensembl. LAGAN21 was used for multiple alignment of the river lamprey, pufferfish, and human NPRL3 genes, and VISTA22 was used for visualizing the results.
Analysis of HB and NPRL3 expression
RNA was isolated from adult L fluviatilis brain and blood and was used for oligo-deoxythymine-primed complementary DNA (cDNA) synthesis. Reverse primers specific for each of the 6 L fluviatilis hb genes were located in the 3′ untranslated region and combined with a common forward primer in exon 2. Each reverse/forward primer combination yielded a cDNA amplicon of unique size. For NPRL3, primers were designed to span exons 2 to 5 and 13 to 14 of the L fluviatilis nprl3 gene. Primer sequences, PCR conditions, and sizes of PCR products are listed in supplemental Information.
Proteomics of L fluviatilis HB proteins
Whole blood and gills of adult river lampreys and gills of ethanol-fixed larvae were lysed by using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, and ∼10 μg of protein was separated on 12.5% SDS-PAGE. The area in the 15-20 kD range was cut into 2-mm slices and used for analysis on an LTQ-Orbitrap mass spectrometer (Thermo Fischer Scientific, Waltham, MA). Details are described in supplemental Materials and methods.
DNase I hypersensitive site mapping
Nuclei were isolated from freshly frozen blood and liver samples obtained from adult river lampreys. Aliquots were treated with increasing amounts of DNase I.23,24 DNA from the DNase I treatment series was digested with NcoI or PvuII, size-fractionated on 0.7% agarose gels, and subjected to Southern blot analysis. Other details are described in supplemental Materials and methods.
ATAC sequencing
Blood was collected from adult P marinus individuals and a 70-mm P marinus ammocete. Blood from adults and ammocetes was pelleted by centrifugation and washed several times with phosphate-buffered saline. Erythrocyte concentration was determined by using a hemocytometer, and ∼50 000 cells were then processed for assay for transposase-accessible chromatin (ATAC) with high-throughput sequencing (ATAC-seq).25 Details of ATAC-seq, including bioinformatics, are described in supplemental Materials and methods.
Vector construction for enhancer reporter assays in sea lamprey and zebrafish embryos
The zebrafish MCS-R2 hb enhancer, intron 5 of the L fluviatilis nprl3 gene, or intron 7 of the L fluviatilis nprl3 gene was inserted into the Hugo's lamprey construct green fluorescent protein (HLC GFP) reporter vector,26 along with the L fluviatilis hb2 promoter. Constructs were sequenced to confirm that they carried the correct inserts. ISec-I meganuclease-mediated transgenesis was performed in P marinus embryos as described previously26-28 in the laboratory of Marianne Bronner (California Institute of Technology, Pasadena, CA). Conventional zebrafish transgenesis was conducted by using the Tol2 transposase system.29 Embryos were imaged at ×60 magnification with a Zeiss Axiocam MRm camera and AxioVision Rel 4.6 software (Zeiss, Oberkochen, Germany). Movies were compiled using ImageJ. Other details are described in supplemental Materials and methods.
Results
Isolation and ab initio sequencing of a river lamprey hb locus
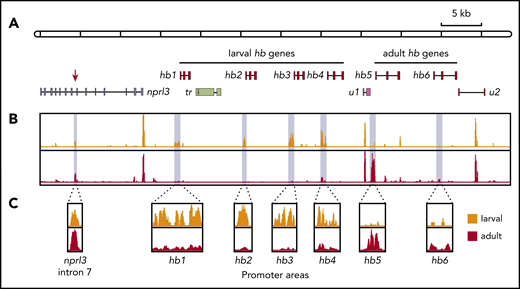
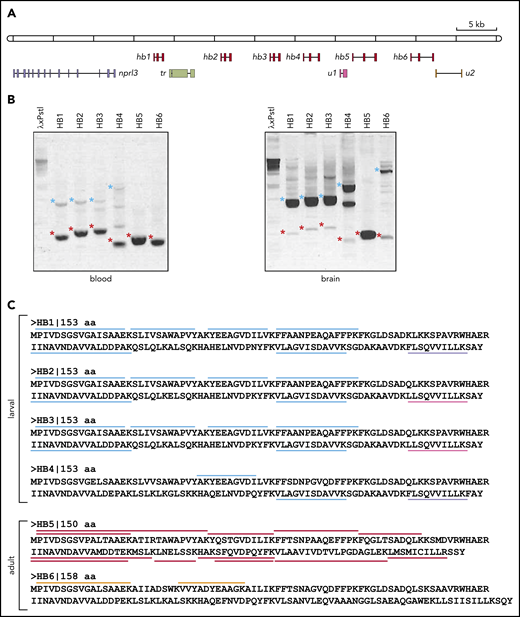
Arrayed filters of an L fluviatilis cosmid library were screened with a 1.2-kb PCR fragment of L fluviatilis genomic DNA. The PCR primers were designed by aligning larval or adult HB cDNA sequences of L zanandreai.30 Positive cosmids were subjected to restriction mapping and Southern blot analysis according to standard procedures.31 Two partially overlapping cosmids (99E08 and 109N16) were selected for analysis by Sanger sequencing of ∼2-kb fragments. Because the genome of L fluviatilis is rich in repetitive sequences, it was challenging to assemble a consensus sequence. So we confirmed the assembly using long-read single-molecule sequencing (Nanopore sequencing; supplemental Figure 1A). GenScan analysis17 of the final assembly identified 10 potential protein-coding genes, including 6 hb genes (hb1 to hb6). Manual curation confirmed their typical 3-exon/2-intron structures with splice donor/acceptor sites at positions conserved in all vertebrate HB genes studied to date,3 and consensus polyadenylation sites (5′-AATAAA-3′) following the third exon. These 6 hb genes were flanked by an apparent ortholog of gnathostome NPRL3 (Figure 1A).
Analysis of the hb genes in the nprl3-linked L fluviatilis hb locus. (A) Schematic drawing (to scale) of the L fluviatilis hb locus. Exons of hb genes are shown as red boxes. To indicate the direction of transcription, gene names are positioned next to the first exon. (B) Expression of the L fluviatilis hb genes assessed by RT-PCR. Red asterisks indicate fragments of expected size for cDNA amplicons; blue asterisks indicate fragments of expected size for genomic amplicons. (C) Proteomic analysis of L fluviatilis HB proteins in larvae and adults. Peptides identified by mass spectrometry are indicated by colored bars: light blue, peptides unique to HB1-HB4; lavender, peptides unique to HB1 and HB4; magenta, peptides unique to HB2 and HB3; red, peptides unique to HB5; orange, peptides unique to HB6. tr, transposon; u1 and u2, predicted genes.
Analysis of the hb genes in the nprl3-linked L fluviatilis hb locus. (A) Schematic drawing (to scale) of the L fluviatilis hb locus. Exons of hb genes are shown as red boxes. To indicate the direction of transcription, gene names are positioned next to the first exon. (B) Expression of the L fluviatilis hb genes assessed by RT-PCR. Red asterisks indicate fragments of expected size for cDNA amplicons; blue asterisks indicate fragments of expected size for genomic amplicons. (C) Proteomic analysis of L fluviatilis HB proteins in larvae and adults. Peptides identified by mass spectrometry are indicated by colored bars: light blue, peptides unique to HB1-HB4; lavender, peptides unique to HB1 and HB4; magenta, peptides unique to HB2 and HB3; red, peptides unique to HB5; orange, peptides unique to HB6. tr, transposon; u1 and u2, predicted genes.
Genomic structure of the river lamprey nprl3-linked hb locus and analysis of gene expression
Our analysis of the genomic structure of the L fluviatilis nprl3-linked hb locus revealed that, remarkably, all 6 hb genes were oriented in the same transcriptional direction (from left to right in Figure 1A). Such an arrangement is similar to that found in the human NPRL3-linked HBA locus.3 We found evidence for recent duplications leading to the L fluviatilis hb1, hb2, and hb3 genes (supplemental Figure 1A-B; supplemental Table 1). Using reverse transcription PCR (RT-PCR), we observed erythroid-specific expression of the hb5 and hb6 genes in adult L fluviatilis blood (Figure 1B; supplemental Figure 1C); expression of hb5 seemed to be the most abundant. In addition, expression of hb1, hb2, hb3, and hb4 was also detected at low levels. To further substantiate these observations, we isolated and analyzed proteins from fresh adult blood and gills, and from the gills of ethanol-fixed larvae (supplemental Figure 2). In the larval samples, we found peptides mapping to HB1-HB4. Most peptides mapped to all 4 proteins, but some mapped specifically to HB1/HB4 or HB2/HB3 (Figure 1C). In the adult blood samples, peptides uniquely mapping to HB5 were abundant; 2 peptides uniquely mapping to HB6 were also detected (Figure 1C). Differential expression of HB genes during development has been universally observed in vertebrates,3 including lampreys.30,32,33 We conclude that HB1 to HB4 are larval and HB5 and HB6 are adult HBs. Furthermore, the results show that all 6 hb genes in this locus are active. These data are largely consistent with a previous survey of HB messenger RNA (mRNA) expression at different developmental stages of the sea lamprey P marinus33 ; HB6 (aHB11 in sea lamprey; see supplemental Information) was described as an embryonic HB in that study. Of note, expression of all 6 hb genes is detectable in adult blood by sensitive RT-PCR assays (Figure 1B); such assays may therefore detect expression of the hb6 gene at embryonic stages as observed by Rohlfing et al.33 Multiple sequence alignments of the 6 lamprey HB proteins and gnathostome globin proteins selected from bony fish (pufferfish and zebrafish), an amphibian (African clawed frog), birds (chicken and zebra finch), and mammals (human and mouse) were used to analyze their phylogenetic relationships. Consistent with previous analyses,8,12,13 we observed that the 6 lamprey HB proteins form a sister group with gnathostome cytoglobin proteins (CYGB), which do not have an oxygen transport function,34 separate from the clades with gnathostome HB and MB proteins (supplemental Figure 3).
Analysis of the nprl3 gene in the river lamprey hb locus
To obtain further insight into the evolutionary origin of extant jawed vertebrate HB loci, we turned our attention to the other genes identified by GenScan in the nprl3-linked L fluviatilis hb locus (Figure 1A). A large predicted peptide of 821 amino acids was derived from a retrotransposon-like element (tr), and was not further considered. Two small predicted peptides of 84 and 74 amino acids (u1 and u2) did not resemble any currently known proteins and were most likely false positives of the GenScan analysis. A predicted peptide of 632 amino acids was highly homologous to the NPRL3 gene linked to the human HBA locus.14 Of note, linkage of an nprl3 homolog to hb genes has been reported for the sea lamprey and the arctic lamprey Lethenteron camtschaticum,13 indicating that this a common feature of cyclostomes (see supplemental Information). The river lamprey nprl3 homolog is actively expressed, as revealed by RT-PCR using primer pairs spanning exons 2 to 5 and 13 to 14 (Figure 2A-B). After manual curation using human NPRL3 as a reference, the river lamprey nprl3 gene is predicted to encode a polypeptide of 581 amino acids, which displays a remarkable homology (69% identity and 80% similarity) to the human NPRL3 protein. A phylogenetic tree of mammalian (human, mouse), bird (chicken, zebra finch), bony fish (pufferfish, zebrafish), cyclostome (river lamprey, sea lamprey), and insect (fruit fly) NPRL3 further illustrates the orthologous relationships between the proteins (Figure 2C). Alignment of pufferfish and human NPRL3 genes to the L fluviatilis hb locus showed that, with the exception of exons 12 and 13 which have merged into a single exon 12 in the L fluviatilis gene, the exon-intron structures of all 3 NPRL3 genes are identical (Figure 2D). Finally, the river lamprey nprl3 gene is located upstream of the larval hb1 gene and transcribed in the direction opposite to that of the hb genes (from right to left in Figure 1A). This structural arrangement is common to the NPRL3-linked HB clusters in most jawed vertebrates studied to date.1,3,14,15,23
Analysis of the L fluviatilis nprl3 gene. (A) Structure of the L fluviatilis nprl3 gene. The predicted exons (green, untranslated regions; purple, coding regions) are numbered. Primers used for RT-PCR are shown, with expected fragment sizes in base pairs (genomic/cDNA). (B) Expression of L fluviatilis NPRL3 mRNA assessed by RT-PCR, using primers shown in panel A, amplifying exons 2 to 5 or 13 to 14. Purple asterisks indicate fragments of expected size for cDNA amplicons; blue asterisk indicates fragment of expected size for genomic amplicon. Bacteriophage λ DNA digested with Pst1 was used as marker (λxPstI). (C) Phylogenetic relationship of human (Homo sapiens [Hs]), mouse (Mus musculus [Mm]), chicken (Gallus gallus [Gg]), zebra finch (Taeniopygia guttata [Tg]), pufferfish (Tetraodon nigroviridis [Tn]), river lamprey (L fluviatilis [Lf]), sea lamprey (Pmarinus [Pm]), fruit fly (Drosophila melanogaster [Dm]), and zebrafish (Danio rerio [Dr]), and NPRL3 proteins inferred by using the maximum likelihood method. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Sizes of proteins are indicated as number of amino acids (aa). (D) VISTA plot displaying alignments of the L fluviatilis, T nigroviridis, and H sapiens NPRL3 genes. Note that the first (noncoding) exon of L fluviatilis nprl3 is not included in the drawing. UTR, untranslated region.
Analysis of the L fluviatilis nprl3 gene. (A) Structure of the L fluviatilis nprl3 gene. The predicted exons (green, untranslated regions; purple, coding regions) are numbered. Primers used for RT-PCR are shown, with expected fragment sizes in base pairs (genomic/cDNA). (B) Expression of L fluviatilis NPRL3 mRNA assessed by RT-PCR, using primers shown in panel A, amplifying exons 2 to 5 or 13 to 14. Purple asterisks indicate fragments of expected size for cDNA amplicons; blue asterisk indicates fragment of expected size for genomic amplicon. Bacteriophage λ DNA digested with Pst1 was used as marker (λxPstI). (C) Phylogenetic relationship of human (Homo sapiens [Hs]), mouse (Mus musculus [Mm]), chicken (Gallus gallus [Gg]), zebra finch (Taeniopygia guttata [Tg]), pufferfish (Tetraodon nigroviridis [Tn]), river lamprey (L fluviatilis [Lf]), sea lamprey (Pmarinus [Pm]), fruit fly (Drosophila melanogaster [Dm]), and zebrafish (Danio rerio [Dr]), and NPRL3 proteins inferred by using the maximum likelihood method. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Sizes of proteins are indicated as number of amino acids (aa). (D) VISTA plot displaying alignments of the L fluviatilis, T nigroviridis, and H sapiens NPRL3 genes. Note that the first (noncoding) exon of L fluviatilis nprl3 is not included in the drawing. UTR, untranslated region.
Chromatin accessibility of the lamprey nprl3-linked hb locus
In jawed vertebrates, distal erythroid regulatory elements of the HB genes are invariably present in the introns of the linked NPRL3 gene.14,15 Where tested, the 2 strongest enhancers are MCS-R1 and MCS-R2 which, in the mouse, contribute to ∼40% and ∼50% of Hba expression, respectively.16 MCS-R1 is located in intron 7, and MCS-R2 is located in intron 5 of the mouse Nprl3 gene. Such elements display strong sensitivity to DNase I digestion in erythroid cells.35 We therefore initially used Southern blotting to map DNase I hypersensitive sites (HSs) in the L fluviatilis nprl3 gene in liver and erythroid cells. Nuclei isolated from adult L fluviatilis erythrocytes and liver were digested with increasing amounts of DNase I. After purification, the DNA samples were digested with NcoI or PvuII and subjected to Southern blot analysis. The locations of restriction sites and the probes used are shown in supplemental Figure 4A; owing to the virtually ubiquitous presence of simple and complex repeats in cyclostome genomes,10 it was not possible to design probes abutting the ends of the restriction fragments.
We found a DNase I HS in intron 7, which was erythroid-specific since it was absent in liver nuclei (eHS; supplemental Figure 4). These data suggest that the eHS corresponds to jawed vertebrate MCS-R1. To further investigate this, we used ATAC-seq25 to assess chromatin accessibility throughout the entire hb locus. These experiments were performed using erythrocytes isolated from sea lamprey larvae and adults. Consistent with the Southern blot analysis, we observed an ATAC site in intron 7 of nprl3 that was present in samples with larval and adult origin (Figure 3). In addition, the promoter regions of the hb1-hb4 genes displayed an extensive open chromatin conformation in larval erythrocytes. In contrast, in adult erythrocytes, the promoter area of the hb5 gene was the most highly accessible (Figure 3C). In agreement with the RT-PCR and proteomics data (Figure 1B-C), the accessibility of the hb6 promoter area was unremarkable in both larval and adult erythrocytes, supporting the notion that hb6 encodes a minor HB. Collectively, we conclude that the chromatin landscapes of the lamprey hb locus at these 2 developmental time points are remarkably similar to those observed for mammalian HBA loci.16
Chromatin accessibility of the nprl3-linked hb locus mapped by ATAC-seq. (A) Schematic drawing of the nprl3-linked hb locus. Intron 7 of the nprl3 gene is marked by a red arrow. Other details are the same as in Figure 1A. (B) ATAC-seq analysis of larval (orange) and adult (red) lamprey blood. Light blue shading indicates areas enlarged in panel C.
Chromatin accessibility of the nprl3-linked hb locus mapped by ATAC-seq. (A) Schematic drawing of the nprl3-linked hb locus. Intron 7 of the nprl3 gene is marked by a red arrow. Other details are the same as in Figure 1A. (B) ATAC-seq analysis of larval (orange) and adult (red) lamprey blood. Light blue shading indicates areas enlarged in panel C.
Functional analysis of putative erythroid enhancers in transgenic lamprey embryos
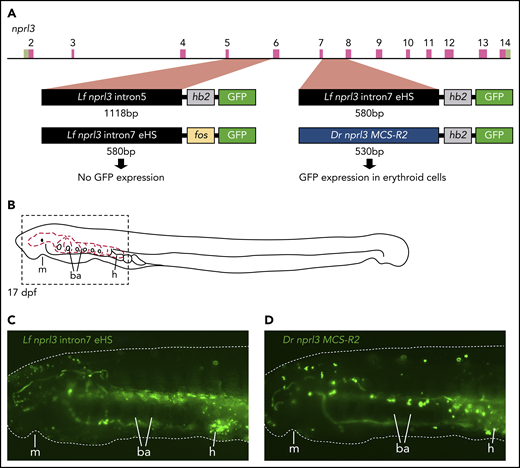
To test the ability of these regions to act as erythroid-specific enhancers, intron 5 or 7 of the L fluviatilis nprl3 gene was linked to the hb2 promoter and cloned into an HLC GFP reporter vector.26 These constructs were used in ISce-I meganuclease-mediated transient transgenesis in the sea lamprey (Figure 4A). By using the L fluviatilis nprl3 intron 7 element, we first noted erythroid-specific GFP expression in the circulation at 12 days postfertilization (dpf), which intensified by 17 dpf (Figure 4B-C; supplemental Movie 1). We observed a similar level of GFP reporter expression in circulating blood cells using the reporter vector containing the zebrafish (Danio rerio) MCS-R2 hb enhancer36 (Figure 4A-D; supplemental Movie 2). In contrast, we did not detect enhancer activity when intron 5 of the L fluviatilis nprl3 gene was used. Finally, a reporter vector using the standard fos promoter downstream of the nprl3 intron 7 region yielded greatly reduced GFP expression in erythroid cells, indicating that activation by the intron 7 enhancer is promoter specific. Thus, the DNase I HS in intron 7 of the L fluviatilis nprl3 gene marks the location of an erythroid-specific enhancer that drives gene expression from the hb2 promoter in a manner similar to that of the MCS-R2 hb enhancer in zebrafish. Inspection of the sequence of L fluviatilis nprl3 intron 7 revealed clustering of several potential binding sites for the well-known erythroid transcription factors GATA1 (GATA box), KLF1 (GC/GT box), NF-E2 (MARE), and TAL1 (E-box) (supplemental Figure 5). This is a hallmark of distal enhancers of HB gene activation in mammals.1,3,14,15,23 We conclude that this erythroid enhancer in river lamprey corresponds to the MCS-R1 element present in jawed vertebrates.
Erythroid-specific enhancer activity of intron7 of L fluviatilis nprl3. (A) Diagram of the L fluviatilis nprl3 gene and GFP reporter vectors used in transgenic enhancer reporter assays in P marinus. (B) Diagram of a 17-dpf P marinus embryo showing the circulatory system (red dashed lines) of the head and branchial arches. Dashed box indicates the region shown in panels C and D. (C) Still from supplemental Movie 1 showing erythroid-specific GFP reporter expression in circulation when intron7 of L fluviatilis nprl3 is used to drive GFP expression from the L fluviatilis hb2 promoter. (D) Still from supplemental Movie 2 showing erythroid-specific GFP reporter expression in circulation when the zebrafish MCS-R2 hb enhancer is used to drive GFP expression from the L fluviatilis hb2 promoter. Dotted white line in panels C and D outlines the embryo; original magnification, ×60. ba, branchial arches; dpf, days postfertilization; Dr, Danio rerio; h, heart; Lf, L fluviatilis; m, mouth.
Erythroid-specific enhancer activity of intron7 of L fluviatilis nprl3. (A) Diagram of the L fluviatilis nprl3 gene and GFP reporter vectors used in transgenic enhancer reporter assays in P marinus. (B) Diagram of a 17-dpf P marinus embryo showing the circulatory system (red dashed lines) of the head and branchial arches. Dashed box indicates the region shown in panels C and D. (C) Still from supplemental Movie 1 showing erythroid-specific GFP reporter expression in circulation when intron7 of L fluviatilis nprl3 is used to drive GFP expression from the L fluviatilis hb2 promoter. (D) Still from supplemental Movie 2 showing erythroid-specific GFP reporter expression in circulation when the zebrafish MCS-R2 hb enhancer is used to drive GFP expression from the L fluviatilis hb2 promoter. Dotted white line in panels C and D outlines the embryo; original magnification, ×60. ba, branchial arches; dpf, days postfertilization; Dr, Danio rerio; h, heart; Lf, L fluviatilis; m, mouth.
Functional analysis of putative erythroid enhancers in transgenic zebrafish embryos
Because the zebrafish MCS-R2 hb enhancer showed activity in the lamprey, we sought to determine whether the reciprocal experiment would result in reporter activity in zebrafish erythroid cells. To investigate this, we used Tol2-mediated transient transgenesis of the HLC GFP reporter vectors. The zebrafish MCS-R2 hb enhancer drove GFP reporter expression from the L fluviatilis hb2 promoter in the blood islands at 30 hours postfertilization (hpf) and in circulating erythroid cells at 50 hpf (supplemental Movie 3). In contrast, we did not observe any reporter activity above background expression when intron 5 or intron 7 of the L fluviatilis nprl3 gene was used to drive GFP expression. These observations show that, although the zebrafish MCS-R2 enhancer has retained properties that allow it to be recognized by the lamprey transcriptional machinery in a tissue-specific manner, the L fluviatilis MCS-R1 enhancer is not functional in the zebrafish.
Discussion
HB gene clusters have been studied intensively as models for tissue-specific, high-level, developmentally regulated gene expression. The discovery and detailed molecular characterization of distal regulatory elements that activate HB expression in erythroid cells have been instrumental for developing current gene therapy vectors for the treating patients with β-thalassemia or sickle cell disease.37-39 Although the evolutionary origin of the HB genes and proteins can be traced by multispecies sequence alignments followed by phylogenetic analyses,3,4,8,13 the distal regulatory elements often display poor sequence conservation even between relatively closely related species such as mice and humans.1,14,15 Identifying these elements therefore requires a different experimental approach that may include localization of clustered binding sites for specific transcription factors such as GATA1, KLF1, NF-E2, and TAL1, mapping of local chromatin properties such as histone modifications and DNAse I/ATAC HSs, and functional analysis by linking putative distal regulatory elements to reporter genes in stable transgenesis assays.1 This approach has revealed the general molecular principles underlying developmentally regulated HB expression, addressing, for instance, the silencing mechanism of the fetal HBG1/2 genes40-42 and the interplay of multiple distal regulatory elements in HBA1/2 gene activation.16 Mammalian HBA genes are invariably linked to the ubiquitously expressed NPRL3 gene. Major erythroid-specific distal regulatory elements are located in intron 5 and intron 7 of NPRL3. Bony fish also contain an nprl3-linked hb locus,14,36 and in zebrafish, the presence of a distal regulatory element in nprl3 intron 5 has been demonstrated by biochemical and transgenic analysis.36
On the basis of comparative analysis of pufferfish and human globin loci, some of us previously proposed that the linkage of HB genes to NPRL3 occurred after the diversification of the monomeric HBs of jawless vertebrates into the tetrameric HBs of jawed vertebrates.23 However, the striking structural and functional similarities between the L fluviatilis hb and mammalian HBA loci reported here provide unambiguous support for an ancient, common evolutionary origin of NPRL3-linked HB loci. These similarities can be summarized as follows. First, the lamprey hb locus contains multiple hb genes all oriented in the same transcriptional direction and arranged in order of developmental expression. Second, the lamprey nprl3 gene is located upstream and in the opposite transcriptional direction to the larval hb genes. We note that, as in the human genome, the lamprey genome contains only 1 nprl3 gene. Third, the chromatin accessibility landscape of the lamprey hb locus displays an ATAC site at nprl3 intron 7 in larval and adult erythroid cells. In contrast, the hb gene promoters display ATAC sites only when the genes are active (ie, the larval genes in larval cells and the adult hb5 gene in adult cells). Finally, functional analysis shows that the lamprey nprl3 intron 7 ATAC site corresponds to the mammalian MCS-R1 erythroid distal enhancer element located in NPRL3 intron 7. Thus, our data strongly support an ancient common evolutionary origin of NPRL3-linked HB loci in jawless and jawed vertebrates. Consequently, our previous model in which linkage of HB genes to NPRL3 was proposed to occur after the diversification of jawless and jawed vertebrates23 needs to be redrawn.
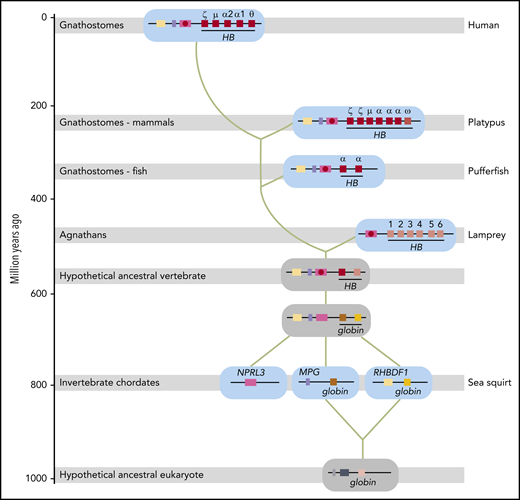
An updated model for the evolutionary origin of the human HBA locus, taking these and other recent observations13 into account, is presented in Figure 5. Of note, in Tunicata and Cephalochordata, which represent Chordata more primitive than Vertebrata, no linkage between nprl3 and globin genes is observed.43,44 The globins do not have respiratory functions in these organisms. In the sea squirt Ciona intestinalis, a tunicate, globin genes are linked to mpg (on chromosome 3) and rhbdf1 (on chromosome 1)44 (Figure 5). MPG and RHBDF1 are hallmark genes of jawed vertebrate NPRL3-linked HB loci.1 This suggests that the coupling between MPG, RHBDF1, NPRL3, and multiple globin genes was first established in the vertebrate lineage (Figure 5), providing a platform to develop erythroid-specific expression via the enhancers located in NPRL3 introns and oxygen transport via adaptations of the globin proteins.44 Despite this common evolutionary origin, comprehensive phylogenetic analysis of globin proteins supports an independent origin of the oxygen transport function of HBs in extant jawless and jawed vertebrates.8,13 Our model provides an explanation for this enigma. We propose that the different globin genes present in the ancestral NPRL3-linked multiglobin gene locus acquired oxygen transport functionality and were recruited by the NPRL3-linked enhancer to drive erythroid-specific expression (Figure 5). Jawless and jawed vertebrates kept different HB genes from this ancestral locus, resulting in the NPRL3-linked HB loci of extant representatives of these 2 vertebrate classes.
Model for the evolutionary origin of the human HBA locus. Based on the results reported in this article, our previous model for the evolutionary origin of the human HBA locus23 has been revised. Genes are color-coded and indicated by gene symbols or names at first appearance. Shaded gray bars indicate when Agnathans (invertebrate chordates) and Gnathostomes (fish, mammals) first appeared in geologic time. Extant representative species are indicated on the right (human, platypus, pufferfish, lamprey, and sea squirt). Loci from these species are shown on a cyan background; inferred loci are shown on a gray background. Green lines indicate the trajectory of evolution of the human HBA locus. Distal erythroid enhancers in introns of the NPRL3 gene are indicated by a red circle. Gene symbols: HB hemoglobin (encoding a globin with oxygen transport function); MPG, N-methylpurine DNA glycosylase; NPRL3, nitrogen permease regulator-like 3 (GATOR1 complex subunit); RHBDF1, rhomboid 5 homolog 1. The evolutionary time scale is based on Burmester et al.45
Model for the evolutionary origin of the human HBA locus. Based on the results reported in this article, our previous model for the evolutionary origin of the human HBA locus23 has been revised. Genes are color-coded and indicated by gene symbols or names at first appearance. Shaded gray bars indicate when Agnathans (invertebrate chordates) and Gnathostomes (fish, mammals) first appeared in geologic time. Extant representative species are indicated on the right (human, platypus, pufferfish, lamprey, and sea squirt). Loci from these species are shown on a cyan background; inferred loci are shown on a gray background. Green lines indicate the trajectory of evolution of the human HBA locus. Distal erythroid enhancers in introns of the NPRL3 gene are indicated by a red circle. Gene symbols: HB hemoglobin (encoding a globin with oxygen transport function); MPG, N-methylpurine DNA glycosylase; NPRL3, nitrogen permease regulator-like 3 (GATOR1 complex subunit); RHBDF1, rhomboid 5 homolog 1. The evolutionary time scale is based on Burmester et al.45
In summary, we show that comparison of the genomic environment in jawless and jawed vertebrates, including determination of tissue-specific chromatin accessibility, functional characterization of distal cis-acting elements, and analysis of developmentally regulated gene expression, provides critical information about the evolutionary origin of multigene loci.
The nprl3-linked L fluviatilis hb locus sequence has been submitted to the NCBI nucleotide database (accession number MK495953). Oxford Nanopore sequencing data are available from R.J.G. at richard.gibbons@imm.ox.ac.uk; proteomics data are available from J.A.A.D. at j.demmers@erasmusmc.nl. ATAC-seq data have been deposited in the European Nucleotide Archive (https://www.ebi.ac.uk/ena; accession number PRJEB31091).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Jim Hughes and Maria Suciu for help with the initial chromatin accessibility experiments and the reviewers for their thoughtful comments.
This work was supported by the Netherlands Genomics Initiative, the Landsteiner Foundation for Blood Transfusion Research (1040 and 1627), and the Netherlands Scientific Organization ZonMw (DN 82-301, 912-07-019, and 40-00812-98-12128). D.H. was supported by a European Molecular Biology Organization Short-Term Fellowship (ASTF 337-2014). Research in the laboratory of D.R.H. was supported by the Medical Research Council (United Kingdom).
The nanopore sequencing was undertaken as part of the MinION Early Access Program, and the MinION sequencing apparatus and flow cells were supplied by Oxford Nanopore Technologies without charge.
Authorship
Contribution: M.M., D.H., R.J.G., L.I.Z., F.G., T.S.-S., D.R.H., and S.P. designed the experiments; M.M., N.G., D.H., J.-F.C., E.M., M.S., C.A.F., and J.J.G. performed the experiments; M.M., N.G., D.H., J.A.A.D., J.-F.C., J.H., S.T., E.M., D.R.H., and S.P. analyzed the data; M.M., N.G., D.H., R.J.G., L.I.Z., F.G., E.M., T.S.-S., D.R.H., and S.P. interpreted the results; M.M., D.H., E.M., D.R.H., and S.P. wrote the manuscript; and all authors reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.M. is Temasek Polytechnic, School of Applied Science, Singapore.
The current affiliation for J.H. is Guangdong General Hospital, School of Medicine, South China University of Technology, Guangzhou, Guangdong, China.
Correspondence: Sjaak Philipsen, Department of Cell Biology, Ee1071b, Erasmus MC, PO Box 2040, 3000 CA, Rotterdam, The Netherlands; e-mail: j.philipsen@erasmusmc.nl; and Douglas R. Higgs, The Weatherall Institute of Molecular Medicine, University of Oxford, MRC Molecular Haematology Unit, Headington, Oxford OX3 9DS, United Kingdom; e-mail: doug.higgs@imm.ox.ac.uk.
REFERENCES
Author notes
M.M., N.G., and D.H. contributed equally to this study.

![Analysis of the L fluviatilis nprl3 gene. (A) Structure of the L fluviatilis nprl3 gene. The predicted exons (green, untranslated regions; purple, coding regions) are numbered. Primers used for RT-PCR are shown, with expected fragment sizes in base pairs (genomic/cDNA). (B) Expression of L fluviatilis NPRL3 mRNA assessed by RT-PCR, using primers shown in panel A, amplifying exons 2 to 5 or 13 to 14. Purple asterisks indicate fragments of expected size for cDNA amplicons; blue asterisk indicates fragment of expected size for genomic amplicon. Bacteriophage λ DNA digested with Pst1 was used as marker (λxPstI). (C) Phylogenetic relationship of human (Homo sapiens [Hs]), mouse (Mus musculus [Mm]), chicken (Gallus gallus [Gg]), zebra finch (Taeniopygia guttata [Tg]), pufferfish (Tetraodon nigroviridis [Tn]), river lamprey (L fluviatilis [Lf]), sea lamprey (Pmarinus [Pm]), fruit fly (Drosophila melanogaster [Dm]), and zebrafish (Danio rerio [Dr]), and NPRL3 proteins inferred by using the maximum likelihood method. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Sizes of proteins are indicated as number of amino acids (aa). (D) VISTA plot displaying alignments of the L fluviatilis, T nigroviridis, and H sapiens NPRL3 genes. Note that the first (noncoding) exon of L fluviatilis nprl3 is not included in the drawing. UTR, untranslated region.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/136/3/10.1182_blood.2020004826/1/m_bloodbld2020004826f2.png?Expires=1765889471&Signature=VQBeQSExzE6Vk3UQs83cAjI2om-rtFYAbFgqCgN~nvD4gUV5AXGo5O-yxXUtE7wOxK50VQVATOFiviqb8n7fPu2W5leWb6oU4YHCAaD0Gvt3VBIsRX1eb6oYAYoutZrcBq68li4WwFloSmTFt4vptOp3iZK5-0-q4OtExgKb2RdvmBgTzdmLx0Kur1LOOiTya0kohE1GDlN1zr6o8UwCNdQgXDU2LYBPGX8bOWqKq8FI~0NZNX-1NUGup~0L~7ybEauDYQexupSm0e5T5FfGEqffJyamnrm~~ULvZrqxS3iQDyFWhtGPujqw6~hKcDZUgfV2O9U1tZZUU3HZl1BGew__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)